













| General Information: |
|
| Name(s) found: |
CE01115
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 448 amino acids |
Gene Ontology: |
|
| Cellular Component: |
pyruvate dehydrogenase complex
[IEA oxoglutarate dehydrogenase complex [IEA extracellular region [IEA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP metabolic process [IEA glycolysis [IEA tricarboxylic acid cycle [IEA pyruvate metabolic process [IEA |
| Molecular Function: |
dihydrolipoyllysine-residue succinyltransferase activity
[IEA protein binding [IEA acyltransferase activity [IEA hormone activity [IEA dihydrolipoyllysine-residue acetyltransferase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMAARLLGTS SRIFKLNKHL HTSKVAFMPV VQFKLSDIGE GIAEVQVKEW YVKEGDTISQ 60
61 FDKVCEVQSD KAAVTISCRY DGIVKKLYHE VDGMARVGQA LIDVEIEGNV EEPEQPKKEA 120
121 ASSSPEAPKS SAPKAPESAH SEGKVLATPA VRRIAIENKI KLAEVRGTGK DGRVLKEDVL 180
181 KFLGQVPADH TSGSTNIRTT HQAPQPSSKS YEPLKEDVAV PIRGYTRAMV KTMTEALKIP 240
241 HFGYNDEINV DSLVKYRAEL KEFAKERHIK LSYMPFFIKA ASLALLEYPS LNSTTDEKME 300
301 NVIHKASHNI CLAMDTPGGL VVPNIKNCEQ RSIFEIAQEL NRLLEAGKKQ QIKREDLIDG 360
361 TFSLSNIGNI GGTYASPVVF PPQVAIGAIG KIEKLPRFDK HDNVIPVNIM KVSWCADHRV 420
421 VDGATMARFS NRWKFYLEHP SAMLAQLK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
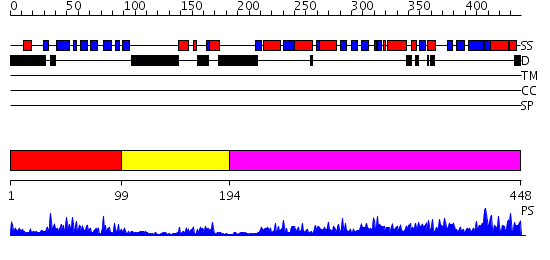
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 18.522879 | Lipoyl domain of the mitochondrial branched-chain alpha-ketoacid dehydrogenase | |
| 2 | View Details | [99..193] | 3.45 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 3 | View Details | [194..448] | 80.69897 | No description for 2ihwA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)