













| General Information: |
|
| Name(s) found: |
umps-1 /
CE00638
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 497 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
'de novo' pyrimidine base biosynthetic process
[IEA positive regulation of growth rate [IMP nucleoside metabolic process [IEA adenine salvage [IEA pyrimidine nucleotide biosynthetic process [IEA pyrimidine base biosynthetic process [IEA |
| Molecular Function: |
adenine phosphoribosyltransferase activity
[IEA orotidine-5'-phosphate decarboxylase activity [IEA transferase activity [IEA orotate phosphoribosyltransferase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSLTDKTRN GALKRNLLRQ MLKASVFKFG EFQLKSGQIS PIYIDLRECF GHPGLLMLIS 60
61 EAISKQVEIS EVQYAGVLGI PYAALPYASV AAGNYLKKPL LIVRKEAKSY GTKKLIEGLY 120
121 QPNDRLILIE DVVTTGGSIL DVVKVLHTEN LVASDVFCIL DREQGGRQKL QDAGVTLHSL 180
181 LDMQTVLTFL YSTGAIGDEQ WHGIVQALNL PYTSPTKLEI NSELENLSSL PYVENVRTPL 240
241 AERESLTESA LIKKILGIMR RKKSNLCLAV DYTTVEQCLQ MIELAGPFVL AIKLHADAIT 300
301 DFNEEFTRKL TTMANDMDFI IFEDRKFGDT GNTNLLQLTG AQKIANWADV VTVHAVQGSD 360
361 SIAGVFRKLA KDPTYRLSGV LLIAQLSTKG SLTALEGYTE TAVKIANENR DVISGFITQT 420
421 RVSACSDLLN WTPGVNLDAK SDSAGQQWRG VDEAIEVQQN DIIIVGRGVT SSSEPVQQLK 480
481 RYRQIAWDAL TRSDDSI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
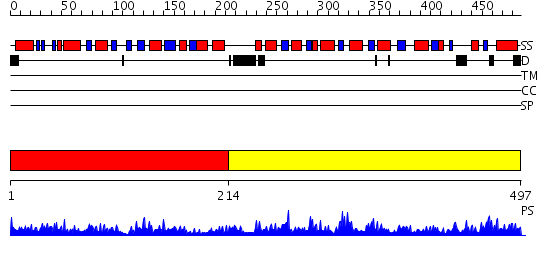
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..213] | 45.522879 | Phosphoribosylpyrophosphate synthetase | |
| 2 | View Details | [214..497] | 73.69897 | No description for 2eawA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)