













| General Information: |
|
| Name(s) found: |
sfc4 /
SPCC16C4.14c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1006 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA transcription factor TFIIIC complex [IDA |
| Biological Process: |
transcription initiation from RNA polymerase III promoter
[IDA regulation of transcription [IC |
| Molecular Function: |
chromatin insulator sequence binding
[IDA RNA polymerase III transcription factor activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIQNGGNSYV DSNMNETQND TTDNFDAEMQ DLNGYISEIV DEARNVSEVD AKFLGDTSAL 60
61 QAEGLWSDEE SDYEGSDDES NFSKTASRTE DDIANEEWEE NLKAVAGFRK VRKGHKGRGR 120
121 VSRADMLPSV EVQQMLSLAN HLFAQEGNFD EAQKLAEEIV RIDNNVIAAW KMLGECHRQR 180
181 GNGRVNIEKC LIAWMAAAHL KPKDHELWFT CAKLSESLEF WDQADYCYNR AVSAKPPNKS 240
241 ELKKYIWNRS VLNKEHGSLK KAAEGFKFLL QSSPYNASIL KNLAEIYIKI HAPREILKQF 300
301 EIAWKYFYQY PAPPIGNDIF DLPTLNLYAE LLLLDHQWSN LIRLINRGVR WFRGRKSESF 360
361 WDEFDDDREW DVDERRREFP NASEEHTNKE AYLLPHLFRT KLGIARLKTG ELPEAELHFS 420
421 VIKNLPPDYA WGMLYDIAKA YMDIERLDLA LEYFVLICNH EPAQNIGLWY NMGVCYLELK 480
481 EYEHAQQCME AILIVDNSNT NALIKLAEIN ELQDNRDAAL EIVTNIFEQR RNINELEREQ 540
541 SQNEDHEKNV GSQLFVGNQK VPQDKWEKRA RISRSKEEAR QFTIWKTEET QRRFHKLDIL 600
601 RQSLKKEENV SESLNEWLAI ASELIDEFVS IKAFFPSEKK ARARAGLLTR RTRYASLNDQ 660
661 LTSMINRLND SLTRTKYGDL DLDTILRTGY FRNVSIDAWY QLFVEFSLRL TKVGSVQQAY 720
721 DVLTTAMGAI LFDQDTIKRQ NLRWCMLACS MYARDPQGAL TPLRWVFTTF QFRQDTYRLF 780
781 SAVLSQGYEC SRAFVDSANQ KFLLRLIKLM DQLMSNSLVS GAATLVKNDD GLATVPTSYD 840
841 PVLVLLYGHI MARNRSWIPA INYYSRAFAI NPDCPITNLS LGLAYLHRAM QRLSDNRHYQ 900
901 ILQGFTFLYR YYDLRVNEGL GEKQEALYNL GKAYHFIGLE HYAVKYYEAV LGLSPMSQGD 960
961 KMTSSESTVS TTYDFGFEAA YNLRLIYICS GNIKLAFQIS SKYLIF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
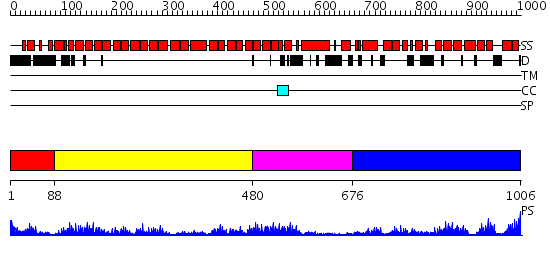
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [88..479] | 45.69897 | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. | |
| 3 | View Details | [480..675] | 4.0 | Crystal Structure of the Hypothetical protein VPA1032 from Vibrio parahaemolyticus, Northeast Structural Genomics Target VpR44 | |
| 4 | View Details | [676..1006] | 22.045757 | No description for 2j9qA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)