













| General Information: |
|
| Name(s) found: |
vps8 /
SPAC17A2.06c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1272 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA CORVET complex [ISS] Golgi apparatus [IDA |
| Biological Process: |
late endosome to vacuole transport
[ISS intracellular protein transport [RCA protein ubiquitination [ISS |
| Molecular Function: |
zinc ion binding
[NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQSKYGRSSL KIKNGDDTRT LRALSFSKDS VNSSSFDNEA ASTTLGISTA IHASNRPFRQ 60
61 DEYSWNDCWM RWGQLRRVSS QLRELRFTMN DQPATVTSIA YQGTLLVAGT SSGHVLLNDW 120
121 RTNDFQILKP GLSSNESVTL PVTSLAISNS KRIVCQGHAG GIIFVWDVSR KPPLQLFTIN 180
181 QHSEDSVLTH LVFNGNSDDV LLSTDHLGKI AVHEFYNLVI NKHCTSWNLD MSKSNSLNLD 240
241 SIIVDTSSIS FEELHITGVK HFSFVVLQSL QSIQIVTLFP KTTLIYEFKY PAGIVSSACH 300
301 SFSSSIVHKN GENTKLHAFF ATAYNNNLRL YSVTFHKGYM SLQLMDRKKF AKAIWKLWWF 360
361 PSSSIIIILF DDMELAFVNS MSLDIIGTST ILDKSLLRWD WYSNSLAAIG VSQTVFPFIS 420
421 SSLCISPRYM FVVNSETVSV GSFLMVSEQL QMLSERYGFF DSIKFGSYCV SNLSIFPKLL 480
481 LEKTSVINLI MALYVKLLKS LARDFPARSP FEATSQKTND YELEVQHLFT WTCELYSENF 540
541 NTVISSFIIA NSLGILPEII EQIIPVFTRV GKVYVVLEAL FDLILSQRFT NPSPQLQHHI 600
601 LNYLNDCKRL QDMDKLIPCI EYQSLDLDFI TKFSRSKGLF DSLCYVSIYA FNDYSIPLVQ 660
661 FLNLLKSDID SPSEETSINV EKGFHFLLYS LTGMKYPLGH SNNINDAYVV IHTLLKVIFS 720
721 VVPLPFDVPV DNSVDFPYLR LFLEKHADDF FRCLEVAFDY PYFSLEKVTI AKKLLDTVNR 780
781 QWILECISQL YEKKKSVSQS YYIFVSHVTA NYSNQLLFSQ SFYQGILDGL CNIDVPDDKR 840
841 EVVEAAIEEL LTVYEHFDIA TTLLSCWDHQ LYQVILHISF KCGRYEDYIE AILALWKQKG 900
901 QKHSFVSESS EALISLVFNN LQTINRFPGH RERYVSTLIS HCGDLFAINH VCFVRNSLKY 960
961 LLDDFTKVIT KTFSIKSIRL QFLSLIIYEI TYEQLSSWYR ERTILSFLEL VSFDQGDVKM1020
1021 LELLRLHPKL VRLQGLMEIL NKNDCIESCI FVYRELAEYK LALSHVSKYI EKSMSVFDLE1080
1081 KSDDMTSRRI ERLCLHLKTV VPLFEQCTKE VSAEDVTNFW LELVFTLLLV YIKLSLKGSI1140
1141 NSQITFKDFQ QELSLCIENL LDSFLSKTNS YKIALNTVLV RICEEASRNE HSDQYLRKLL1200
1201 LKLITDLYIN HDITKECFLL WDMKQFYEIR STLLQNASGV LVEDPKLQKN AKGQYTSKLK1260
1261 VYFSGLIERE DL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
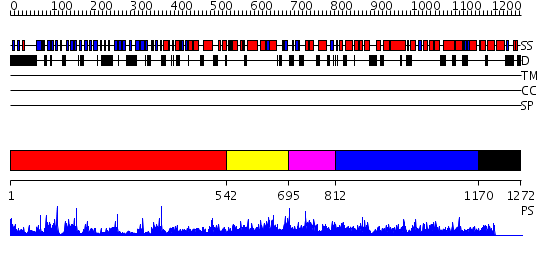
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..541] | 45.39794 | Actin interacting protein 1 | |
| 2 | View Details | [542..694] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [695..811] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [812..1169] | 4.69897 | Clathrin D6 Coat | |
| 5 | View Details | [1170..1272] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.46 |
Source: Reynolds et al. (2008)