













| General Information: |
|
| Name(s) found: |
SPBP8B7.29
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 718 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [ISS cytosol [IDA |
| Biological Process: |
para-aminobenzoic acid biosynthetic process
[ISS glutamine metabolic process [IEA] folic acid biosynthetic process [IEA] |
| Molecular Function: |
oxo-acid-lyase activity
[IEA]
4-amino-4-deoxychorismate synthase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEISNRLQI LLIDCYDSYT FNLYDLLYKA SENACVIVVH WDKMSPDLWE DILQFDAIVV 60
61 GPGPGHPAEY SSILNRIWQL NIPVMGICLG FQSLALYHGA TIERMPNLPW HGRVSSVTTS 120
121 KTFIFDGISA VKGMRYHSLY ANKIPIDSLQ ILAQSDEDNI VMSIKATKFP HFGILYHPES 180
181 VGSSKSLKIF KNFLSLADTP NIQCVNSFSK SANGFSHNLN RYDISPAAFI LKSGSPSLQI 240
241 HSVEIPWVEP LALADCIQKS GNPICFLDSA KKPGRYSILG ILTGPLARII HYEKATNTTE 300
301 IRICKDNSFV RINNDLWSTV ADFMNQHKAI KPDTNLPFYG GIMGIIGYEC SDLSTKSVSN 360
361 ASFPLDFQQT TVDAELAFVD RSFVFDLEIK KLFVQTLTPL NETCSEWWGE LLASTCNTKL 420
421 DNLSCLHSFD GKQNFGLVQS FPKKEVYCES VKACQEHLLA GDSYEMCLTD TTFVSAPPEL 480
481 SDFEMYMRAR SLNPATFAGF VRLNHFTLLC CSPERFLQFR DDRCLFSPIK GTLKREGHMS 540
541 LEEARKKLLN EKDMGELNMI IDLIRNDLHQ LAKKNSVHVP ELYSVEEHSN VYSLLSNIYG 600
601 RIESPITAWD VLSKSFPPGS MTGAPKLRSV RMLEPLEQHG RGIYSGTLGY WDVTGSAEFN 660
661 VIIRSAFKYK ADDYWRIGAG GAVTILSSPE GEYEEMVLKA NSILPAFVNL KNKKRSCK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
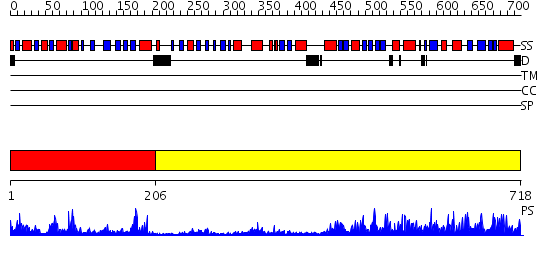
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..205] | 40.69897 | Anthranilate synthase GAT subunit, TrpG | |
| 2 | View Details | [206..718] | 118.0 | P-aminobenzoate synthase component I |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.47 |
Source: Reynolds et al. (2008)