













| General Information: |
|
| Name(s) found: |
SPCC622.10c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 815 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cell tip [IDA exocyst [ISS |
| Biological Process: |
exocytosis
[ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSADEEILAH YGLTSLSQDY WTEKPDFAIA NASVDVQSPT TGVHHERNRS EDILRTGGAT 60
61 SETTKDASAA GPVTPSFAMK RSQTSVQPSM SSQIAIGGMR NPMATRKIAG TYRDAPDNSM 120
121 QQTSTGVHRS SSWLTTDGTH QLLSLMETNF NKFIAAKETI DNVYAQIIST LSHAGPAKTM 180
181 DMCEESISGA LQQLQLFNTE STNEVDISYG ELLVMRECEA LFTYPEKFRL YHKQEKYSLL 240
241 MQEIENAKKL YEKDRERLAK VNPKLVYLVD HAWEIIQATI DGICASIWNQ ILQSQGDFAF 300
301 VVRTLCDLLK LKPNPISGRD PVLYATVSQQ KYLYNLMKER FSYFEKIIKS RQEDFSKLSC 360
361 TRASPMLVWR AVARGCNLDY TLRGRPEVFA GMQLIEWANA CVLKIVREIK EVELYAKEFI 420
421 SGIQQASYPE AAKHYGRDPK QMGVVVEKLN SQCLEMVYSF VGERGERLEM DGAFVLIQLH 480
481 YFQRVADLLP KRIRTVFSMA ATRCIVDTWM LQHAPQSKSA NIEEMFIVDA NENQVLEPEL 540
541 MVVDVLRELN KLYIPPHREE IVREIHHAFY KVLYANARFP SMSVSMANWE SDVWSLASST 600
601 LIDFVQHRNQ HDMKKLASKM TTSLPVFSTS ITRSDVLHFV TRLLNLRSRL VPYQKFLLQT 660
661 FPLNEVSPSL DPRISKSLRN LQDLYKLLAI RDTSFSNTTP SSISKLSIQP PTFQLFSTLE 720
721 TIYLELKDSM PPNEAADWMI ATTSCMLAQF ALHSPYAFFH DIPSISHFLN KCLPPTILDL 780
781 LQQIQRKAEA SSISPTTETA TKTRLKFQFI EMIFS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
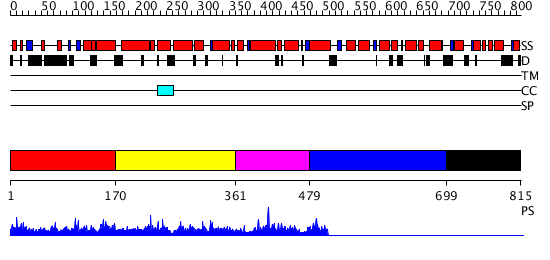
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..169] | 7.084996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [170..360] | 3.082976 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [361..478] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [479..698] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [699..815] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)