













| General Information: |
|
| Name(s) found: |
uba5 /
SPAC323.06c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 500 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
protein neddylation
[ISS |
| Molecular Function: |
ATP binding
[IEA]
NEDD8 activating enzyme activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTSAKMQKY DRQVRLWKAE GQNAIEKSHV CLLYANTVGC EALKNLILPG IGSFAVVDDT 60
61 SVDFSMDGMN FFIQYDQEGK SRARCTASLL QQLNPNVEME YLEMSPEALI DKNIEYFSKF 120
121 SVVLSSNLKE KPLFRLEEYL RSHKIPLLHF NSVGFAGILR ISTHEYTTTQ SQPELPQDLR 180
181 LKNPWPELIN YVKSMDLDNM DSSSLSEIPY IVLIIHVLLK VSPAHAQNSQ EADDCAMFRK 240
241 IMEEYKGKCD SENIEEASSN SWKAFKEYKL PSNVYEVLHD TRCVKIQEDS ESFWIMAHCL 300
301 KMFYDETEFL PLSGLLPDMN CSTQQYVKLQ VIYKEKSEND ILKFKKYVQQ TLKRLNRSVE 360
361 EITDLEIKHF SRNCLNIKVM DFKTMKEEYQ PTSNSAFRIY DTILEKHGKN YKEAFSDTTK 420
421 TISVAQSFLS QIGLEKFFDV VYTAIQELER ADGHELHSIS SFIGGIVAQE TIKLLAQQYL 480
481 PLNNTFVFDG VHSRTETFKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
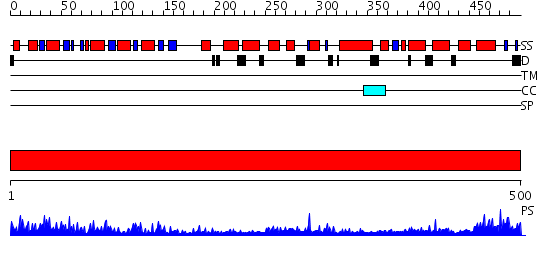
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..500] | 116.0 | Structure of APPBP1-UBA3-Ubc12N26: a unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)