













| General Information: |
|
| Name(s) found: |
SPAC458.03
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 868 amino acids |
Gene Ontology: |
|
| Cellular Component: |
TORC2 complex
[IDA ASTRA complex [IDA cytosol [IDA TORC1 complex [IDA |
| Biological Process: |
regulation of S phase
[IGI DNA replication checkpoint [IGI chromatin remodeling [IPI UV protection [IGI positive regulation of protein amino acid phosphorylation [IGI replication fork arrest [IGI telomere maintenance via telomerase [ISS TOR signaling pathway [TAS |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSFASELSK PIQKPRLKEI LKELQHIGVP SPCNLRLYHE LISVIVPTFW NGKINSEIDF 60
61 LLAKCFSTLL GLKLLCVRLD NFVQEANLQN IHLVTVYLEL LEKAFECIDL DDNCKLIWES 120
121 TVLSDHQKQS LWTEFIFFLG NFKITNSMSA AMLTFKVYDD SKRLKASSMA DYIGVLSSKL 180
181 ACIISKPAVK TPEIYSKLFH YLLHSSNLKA FINPLIPLTQ KFCVQLQKLF ADLTVSDQML 240
241 FLNQLLLEHN TKYPTNFSYS TARDDRITGS LATLLRLNFS STHFLRLIEF YWGVPTNLII 300
301 KRVEVVCSSI SYKESDKKEL EKTVDSLFNI WSNAQFLRSY TLLAQESLTI YLLLFIPKLE 360
361 AKYLNNLSKS LMFSNAISCR LDSLDDDIRM HGMIMAEVIS TYSGVSLSNP LAFDVPIMKT 420
421 TKAKVLKSLS SLKDEFLPIE ILNDESVIAE TSIEKEETNV THLEKPVISK NDDLIRGDDL 480
481 EPYDFPDIDS EDTDDDPTVS RSKTHSPVYV QDLCKMLKDT ESFEKQRVAL ENASKLIKRK 540
541 SAFGTELRDH ADELLQTLIS LQNRFDLMNF DEMQMTAIVE LLLTCLDICG PVICTNLFVS 600
601 DYSMRQKILI LSCISLAASK FNDDDNERLF PSQLLPGNLH DQFYSPTIEK ISDELERKLV 660
661 FPVMSECKDY AEGPKFLQTR YISKQSEIES KKPLPKANRL SLKVDKSLFL LLTSGYMLAS 720
721 RKTAFIEPLF LVYYLKTLGI LFFETFTSKI EGANRMLREY VDVLNEVCRL RSDTPEVNEG 780
781 LLFSFLAILE ISSGRTLANE FGKHLLFFEA YTKFLFESPD QGEKVKSLSA AVLILFENKL 840
841 SEFRTLALEK LLDDSAPLVP ERFGLAGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
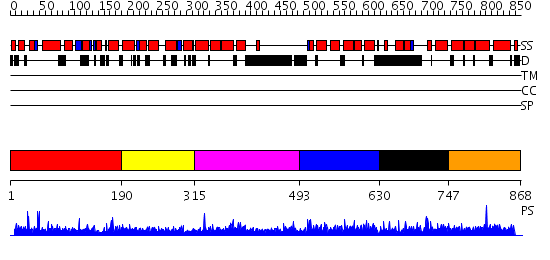
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..189] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [190..314] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [315..492] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [493..629] | 1.044968 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [630..746] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [747..868] | 1.037983 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)