













| General Information: |
|
| Name(s) found: |
gi|7491907
[NCBI NR]
gi|46396277 [NCBI NR] gi|2213548 [NCBI NR] gi|19075333 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 572 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSCPITFHR PSQALALKGI GPTICAKLEK KWNAYCLENN IPISTHNEQN DSHVNANKSS 60
61 SETSSEKPRS VKKPTTRKRK VYVPSYRSGA YSILCALYML NKHEFATKPQ IVTMAQPYCD 120
121 SSFGSATDRN MRYTAWSAMK TLITKNLVYQ TGHPSKYCLT DDGEEVCIRL AKVDDSFQRK 180
181 HTVSNFSVSK SDDHDSSLCQ PPNFVTSINK AGSSSDHGGE LHVTYCPVDH NEVSDGVETD 240
241 IDVDQVDSLT GIHDHHIINN EQLIDLTEQE KKQPNESNLS NLKIETVLFS NCTVFLLIDT 300
301 REIRSPLDRN LIIDKLTNDF GVNCQVRSLE LGDALWVARD MESGQEVVLD FVVERKRYDD 360
361 LVASIKDGRF HEQKARLKKS GIRSVTYILE ESSYDESFTE SIRTAVSNTQ VDQLFHVRHT 420
421 RSLEHSVSLL AEMTKQINLF YEKRKTLAVI PDLSIEAKTY ESLREQLLKI DPSTPYHISY 480
481 HAFSSVLSKS STLTVGDIFI RMLMTIKGIS ASKAIEIQKK YPTFMHLFEA YEKSSSSQER 540
541 NLLLNKTCQG YGFQTIGPAL SAKVASVFFP ES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
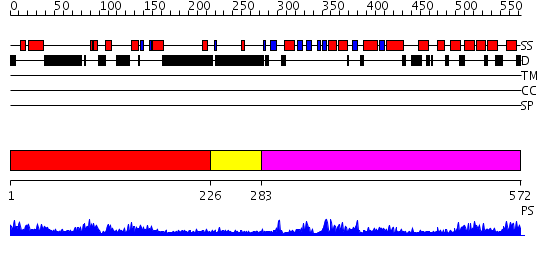
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..225] | 34.0 | Crystal structure of Pyrococcus furiosus Hef helicase domain | |
| 2 | View Details | [226..282] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [283..572] | 38.0 | XPF from Aeropyrum pernix, complex with DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)