













| General Information: |
|
| Name(s) found: |
SPCC594.05c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 424 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Set1C/COMPASS complex
[IDA nucleus [IDA nuclear chromatin [IC cytosol [IDA |
| Biological Process: |
chromatin remodeling
[IC regulation of histone H3-K4 methylation [IMP |
| Molecular Function: |
zinc ion binding
[IEA]
small conjugating protein ligase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELTQENTEE NEKTHVESIV KFEDSNRGTI TDFHIETANN EEEKDANVIL NKSVKMEVEE 60
61 VNGHVDSSST ETDIEMQVIQ QPTIPKKPPV SAHRRGPRKH RGNANSQLNL STADHQRPLY 120
121 CICQKPDDGS WMLGCDGCED WFHGTCVNIP ESYNDLTVQY FCPKCTEEGK GITTWKRKCR 180
181 LRECSNPTRP NSNYCSDKHG VDFFREKVKL STVEPSAIKN LVLFAKKREE FQNLGTVGPT 240
241 LPSQVPPEVV YNFEIEEANR LNAEIVQLNK EKEVASNKKI FLQLIKDSSR RAVLAYKERE 300
301 GIKKDLCGFD SRLLFNQQQM NELWEKVSNG APLSLDMSID PSPESTCFTE KRRCAKHTSW 360
361 QVIFTEDFEL QESNILQKLN MKQTAKDVML EHQKQRCLPG IQYDGYARFC HEQLSPEKMA 420
421 NLLK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
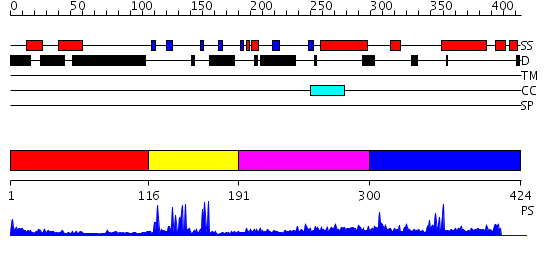
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [116..190] | 14.09691 | Solution structure of PHD domain in PHF8 | |
| 3 | View Details | [191..299] | 3.209996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [300..424] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)