













| General Information: |
|
| Name(s) found: |
SPAC31G5.15
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 980 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA fungal-type vacuole membrane [ISS cytosol [IDA Golgi membrane [ISS |
| Biological Process: |
phosphatidylethanolamine biosynthetic process
[IGI regulation of cell morphogenesis [IGI regulation of cytokinesis [IGI phosphatidylcholine biosynthetic process [ISS |
| Molecular Function: |
phosphatidylserine decarboxylase activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPFTKSCRSA NTLRKGIKNG KLILKIIVND VCCWVLIKVT NKQIDNVESC LSGDESNDSK 60
61 KSSASFYMKL KYGSYRALAD NILSTDENRK EDIAVFDVPL PNGLQIDTFT LCLYRKSKWK 120
121 KQIVGEAYIG IQALLLSTNP DETCKYPVIS PPSGRNKENQ SPSHQICNLS LKWIIYDPED 180
181 ADADSKTLAK AWLQQIKMNQ TSIDPMSNIS KSLKELEVDN VESDLEDSSF IAEPDSSIPP 240
241 SESSVSISTD TGKETPPSKS KKSSNQPYVS IGEGNSDLLG FVFLEIISVS NLPPLKNVFR 300
301 TGFDMDPFVI TAFSKNIFRT KWLRHNLNPV YNEKFLFEVG AFESNYDLVF KVVDHDKMSL 360
361 NDSIAVGSFN VQSIINSSAQ VDPETGLYSF NIETSSPSQD TSSKAEDSPT VQKIADDFSS 420
421 AVGKDLRTDI IEQIIPLTLC CKHDFSTPRD VKLSFKAMFF PIAALRQKFW RVMLAQYGDI 480
481 EDGHIGKLGM YAVLDTLGSN IPNSMVDDIY TELSSKNHDD TSDSITVDEA VICLERLVDL 540
541 VCHQDQQATQ TPQSPSSNEE SGPGTPTQTS DQYEDSEDSR NFPSKLYLVY LSNCPLCLKF 600
601 KLSKVNQQKA TVHLATCASH DWKRVDRLMM TSYVSLNQAQ RRWFSKAFAK VVYGSSKVGS 660
661 TSATTLVQNR QTGQIQEEKM NAYVRIGIRL LYRGIRNRRI EGSKVKKILR SLTLKQGMKY 720
721 DSPISVKEIK PFIRFFDLNM NEVDMPVGGF KTFNEFFYRK LKPGSRPCAF PDNPDILVSP 780
781 ADSRIVAYEC IEKATTYWIK GTEFTVERLL GYSNEAQRFV GGSICISRLA PQDYHRFHSP 840
841 VNGCIGPITK IEGQYYTVNP MAIRSYLDVF GENVRVLIPI DSNEFGKVML VAVGAMMVGS 900
901 TVLTVDEGKI VQRSDELGYF KFGGSTVITL FEPNVTSFDE DLLRNSKTKI ETLVKMGERI 960
961 GQKIDPNKPT DAEDHSKSDS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Unpublished Data |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
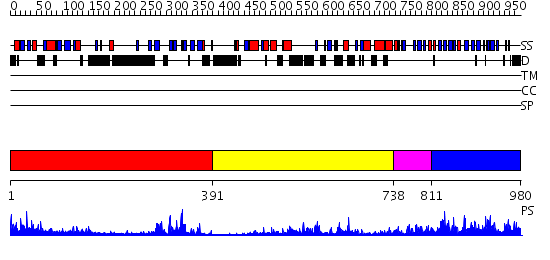
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..390] | 20.69897 | Phosphoinositide-specific phospholipase C, isozyme D1 (PLC-D!); PI-specific phospholipase C isozyme D1 (PLC-D1), C-terminal domain; Phospholipase C isozyme D1 (PLC-D1) | |
| 2 | View Details | [391..737] | 7.69897 | Carboxyl-terminal src kinase (csk) | |
| 3 | View Details | [738..810] | 48.69897 | No description for 2i0eA was found. | |
| 4 | View Details | [811..980] | 5.265982 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)