













| General Information: |
|
| Name(s) found: |
SPBC947.15c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 551 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial inner membrane [ISS |
| Biological Process: |
generation of precursor metabolites and energy
[RCA NADH oxidation [ISS |
| Molecular Function: |
NADH dehydrogenase activity
[ISS FAD binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVSKARLQS VVRLSRTVPY SKTMVRSFHV SCAVKNSGNV PTPRNKSFFS RALEMAEVTS 60
61 SLSMLGAVAL FQSLRRLNNS SPKGKSGVPK KNIVVLGSGW GAVAAIKNLD PSLYNITLVS 120
121 PRDHFLFTPM LPSCTVGTLR LPSITEPIVA LFKGKIDPSN IHQAECTAID TSAKKVTIRG 180
181 TTEANEGKEA VIPYDTLVFA IGAGNQTFGI QGVRDHGCFL KEAGDAKKVF NRIFEILEQV 240
241 RFNKDLSPEE RARLLHITVV GGGPTGMEFA AEMQDFIDND VKDMFPELQK DIHVTLIEAA 300
301 PGVLPMFTKS LITYTENLFK NLNIKIMTKT VVKDVNEKNL IVQKTNPDGS KAMQEIPYGM 360
361 LVWAAGITAR PLTRTLMSSI PEQSGARKGL IVDEFFRVKG VPEMYAVGDC AFSGLPATAQ 420
421 VANQQGAWLA KNLNVEGKKF ALHERIQALE KQLGEKEAPS QVAGLKQQVE QLKLEPFKYH 480
481 HQGALAYVGD EKAIADLKLP FMKKMLPLQG IVGHTFWRLA YLNELISARS QFMVLIDWLK 540
541 TRLFGRYDAK V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
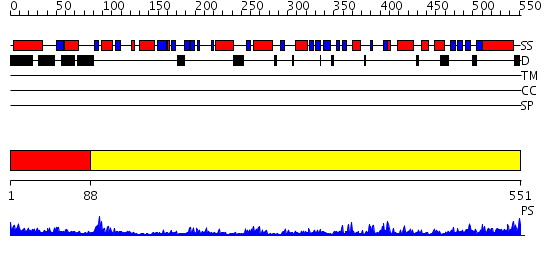
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 37.69897 | The Crystal Structure and Reaction Mechanism of E. coli 2,4-Dienoyl CoA Reductase | |
| 2 | View Details | [88..551] | 69.522879 | Dihydrolipoamide dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.54 |
Source: Reynolds et al. (2008)