













| General Information: |
|
| Name(s) found: |
SPBC2D10.04
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 658 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cell tip [IDA cytoplasm [IDA cytosol [IDA |
| Biological Process: |
regulation of signal transduction
[ISS]
|
| Molecular Function: |
calcium ion binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRGELNIPAL SRFGKSISAS LHHQNGSSRD DNDSNPYENR SSNSGLNRRN SVFGLPSSGL 60
61 SSRLSKPSLS SINNSNNSSS NTGGNVLPNP ALTPVRNMSN KPPLTWSPSS ANLFGTSKVT 120
121 SIIGNNVSDV VPPVIKKALA SSHGGGMSLS IVLLEPVLYL AGFDLNECQI ENPALLRGAL 180
181 VLRVAKPANI RGISLSFTGR SRTEWPEGIP VKGHDTYEDK VIISHNWKFY EPTMKDADAP 240
241 QHGADVARLV GEQLPLPSSA AASLRGYSVF APGEYTYNFD LAIPNCFPES VEAKMGWVRY 300
301 FLEATVERFG TFKSNLNGRT PVQLVRTPSP ASLSSSELIN ISRDWDERLH YELQVSGKSF 360
361 RLGEVVPITF RFLLLDKVRL YKLSISVVES SEYWCRSRKF HRVDPKRRVL LAERSAKHQN 420
421 TDNLFETPDE GDGLSSAVFN FNVALPTCLV KERDRLTFDT TYKYIKVRHR LKALLVLSIE 480
481 NTENPEKRKY FEINIETPVR ILSCRCVKDS TLLPPYESSS QGDNQVLLPC PCRLATTHVE 540
541 PTEVTAFTTQ SVLASSAPSA GRPAAAQISR PAQLFRIPST NPPPFDGDVC PPACNTPPPN 600
601 YDELFDVLSS ISIQDCETDR ANDDTILNNR VRRSGTIREE APHRSLSRTV SRSFEIPR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
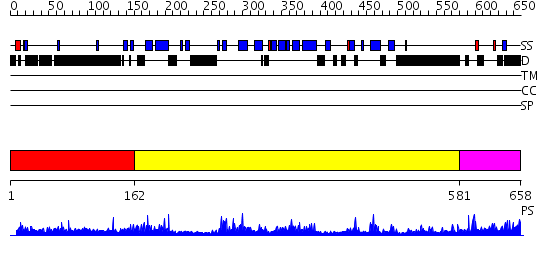
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [162..580] | 69.221849 | Arrestin | |
| 3 | View Details | [581..658] | 2.124996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)