













| General Information: |
|
| Name(s) found: |
SPCC576.05
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1024 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA nucleus [IDA cytosol [IDA nuclear pore [ISS |
| Biological Process: |
mRNA export from nucleus
[ISS protein export from nucleus [ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKRNETGNN RLKRSNNRGK SKKDWKDASV ETTPRETSVD EDNTSVFEDV EAQDSRQKRF 60
61 SSTLEGNRFE ELRSLREKER EVAIQNGLID DPTKPRQLDE AVTFVGTCPD MCPEYEREQR 120
121 EYQNNLERWE INPETGRVDK NLAVKAFHRP AAGNEQALPS DVRPPPVLKK SLDYLVDKIV 180
181 CGPDPLENTH FFVRDRTRSI RQDFTLQNCR DLDAVACHER IARYHILCIH QLCEKKQFSA 240
241 QQEVEQLRKG ILQSLCEFYD DLRKVKIRCP NEPEFRSYAI ITHLRDPDVV RQSQILPIEI 300
301 FDDQRVQLAL RLSALAQKNN ERVGHILPRN TEACPNLYTR FFKLVQSPAV TYLMACLLES 360
361 HFMSIRKGAL KAMRKAFMSA HANFPCGDLK RILHFDTVEQ AASFSRYYGL EVSDDNGELS 420
421 INLNKTAFFN DSKPDFRQLF SQTLVESKLQ NRSFADIING SRYNIDRVSP NTAFSTNIPL 480
481 SLPFANKEPQ PIAGFKKNTP ETSVVSKNLS TFNGKFNVNA PVFTPRSFPT KPFSATDISS 540
541 VQPTNLPNGS TNGTETFIPP VQNSITSNKE AVKPIKNKPK PISFESLSAV GNLIISDSLS 600
601 RIVRQILQNL YTEWVHEKTN LVFATMFRTI FREVLLDGIA SEVYLKSLKK HAISQISVRA 660
661 HHSWVKKQEK MMLEMREKNR QEKYFSVLNS VVKAESSNIT RLPIKRTFYG DTRNLDKASE 720
721 KLRAEHDRTR RLWKPVLMDS LFSNLQKFPV YEDWHLLIFN ASTSSMMKTW LCAKFSLKET 780
781 NKTSFWHSSY NLFNRQYHVD MPDNVSDLPQ TRLCYGACVY NVGLLDEEKR KDLANSDLNS 840
841 SPKLIQGNDS RSAHESSANK LFSFVHDISR LTITKLPLLL IFWSDSNLDM QGITQKYRFL 900
901 ELITSTWSAI SSIHVLTITN DRDMDLEHSL KVLLDNVTVE KSPFAQLEEL EVVRKKREAE 960
961 IEASSKTVKR LASNNKFLTD SNVEGLLEAP TSLENSLVED DKWASLRQKI KAARDLLKKV1020
1021 ETFY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
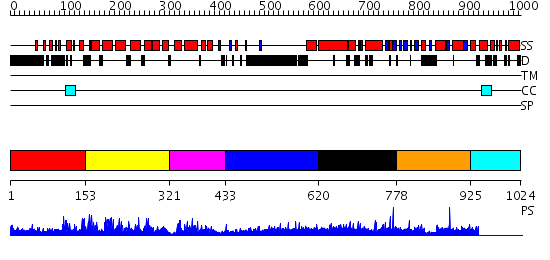
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | 1.106989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [153..320] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [321..432] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [433..619] | 13.235997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [620..777] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [778..924] | 1.00897 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [925..1024] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)