













| General Information: |
|
| Name(s) found: |
SPBC29A3.13
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 359 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [ISS |
| Biological Process: |
DNA damage checkpoint
[IGI chromatin remodeling [ISS regulation of histone H4-K20 methylation [IGI cellular protein localization [IGI |
| Molecular Function: |
protein binding
[IPI methylated histone residue binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNARTNAKR RRLSSKQGGL SISEGKESNI PSVVEESKDN LEQASADDRL NFGDRILVKA 60
61 PGYPWWPALL LRRKETKDSL NTNSSFNVLY KVLFFPDFNF AWVKRNSVKP LLDSEIAKFL 120
121 GSSKRKSKEL IEAYEASKTP PDLKEESSTD EEMDSLSAAE EKPNFLQKRK YHWETSLDES 180
181 DAESISSGSL MSITSISEMY GPTVASTSRS STQLSDQRYP LSSNFDHRGE AKGKGKQPLK 240
241 NPQERGRISP SSPLNDQTKA LMQRLLFFRH KLQKAFLSPD HLIVEEDFYN ASKYLNAISD 300
301 IPFLNYELIT STKLAKVLKR IAFLEHLEND ELYDIRQKCK NLLYSWAMFL PNEPSIKGM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
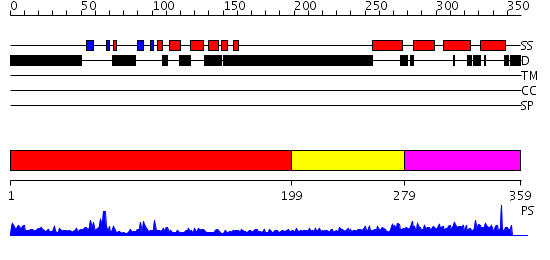
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..198] | 19.69897 | DNA methyltransferase DNMT3B | |
| 2 | View Details | [199..278] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [279..359] | 1.23 | Solution structure of the N-terminal Domain I of mouse transcription elongation factor S-II protein 3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)