













| General Information: |
|
| Name(s) found: |
SPAC27E2.02
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 280 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS cytoplasm [ISS |
| Biological Process: |
regulation of protein amino acid phosphorylation
[ISS]
regulation of translation in response to stress [ISS] |
| Molecular Function: |
translation regulator activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENNEEFQDE LLALESIYPS CLLPISEQSF TYTLSIPDSS VRLNIQFPLD YPNSAPTVLD 60
61 AYGIDKTLAE DVLLSVATGD VCIFSYMDLL KELVDIDAEQ AAAERESKLQ EESDKETPVM 120
121 LNKSHYVAKT PEIQDEPWKP KFDWKESEPI TDRKSTFMAH ATRVYSTEEV REALEDLYMD 180
181 KKVAKANHNM VAYRIISPNG NVIQDNDDDG ESAAGSRMSH LLTMMSAENV FVCVSRWFGG 240
241 VHIGPDRFKH INSSAREAVL LTDAAPSQKK GTEHGKKKKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
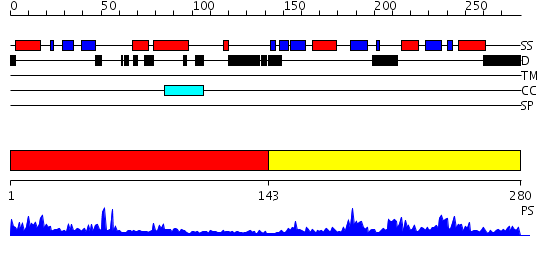
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | 23.522879 | No description for 2ebmA was found. | |
| 2 | View Details | [143..280] | 34.69897 | Crystal structure of a conserved hypothetical protein TT1547 from thermus thermophilus HB8 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)