













| General Information: |
|
| Name(s) found: |
SPBC1734.11
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 407 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
protein targeting to mitochondrion
[ISS protein targeting to ER [ISS] ER-associated protein catabolic process [ISS protein folding [IEA] |
| Molecular Function: |
zinc ion binding
[IEA]
unfolded protein binding [IEA] Hsp70 protein binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKETKLYEV LNVDVTASQA ELKKAYRKLA LKYHPDKNPN AGDKFKEISR AYEILADEEK 60
61 RATYDRFGEE GLQGGGADGG MSADDLFASF FGGGMFGGGM PRGPRKGKDL VHTIKVTLED 120
121 LYRGKTTKLA LQKKVICPKC SGRGGKEGSV KSCASCNGSG VKFITRAMGP MIQRMQMTCP 180
181 DCNGAGETIR DEDRCKECDG AKVISQRKIL TVHVEKGMHN GQKIVFKEEG EQAPGIIPGD 240
241 VIFVIDQKEH PRFKRSGDHL FYEAHVDLLT ALAGGQIVVE HLDDRWLTIP IIPGECIRPN 300
301 ELKVLPGQGM LSQRHHQPGN LYIRFHVDFP EPNFATPEQL ALLEKALPPR KIESAPKNAH 360
361 TEECVLATVD PTEKVRIDNN VDPTTATSMD EDEDEEGGHP GVQCAQQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
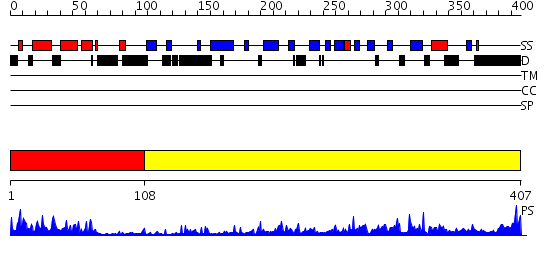
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | 23.30103 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [108..407] | 53.221849 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)