













| General Information: |
|
| Name(s) found: |
SPAC3H8.08c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 563 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS gene-specific transcription from RNA polymerase II promoter [RCA |
| Molecular Function: |
DNA binding
[ISS transcription factor activity [ISS zinc ion binding [ISS specific RNA polymerase II transcription factor activity [RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSPPALKK FRKRSPKSCL ICRRRKVKCD RQQPCSRCKE RNEVCTYADD TIDKMNVGPH 60
61 PSHSENASDS ETTLEVSPDI NPKKNEKFDF YGWRSLFELI KYRKDSDMCS SRPSFSIQAY 120
121 SSYKDNVVVE SLANLLPPFC ISQKIVNLFF KTLNVVCPIY DQETVEKSLN NIESPESFSY 180
181 EDAFTLLPII AATIQLSDLP DVILNFYNSA GITPLESSRL INLKLNEISE QEYKHLCLPD 240
241 KEIIQMLLLR AYATKFRTRI RGVNTDLCRS IHVSTLVTPL FQVTEKIGKN TSDLWFALCE 300
301 IDGLECVLKY RPPFIQHDTY GRLKPLRCFF NDDISYNFHL LLGRLLDCGV SIYKSVHSLT 360
361 VSKFIDKLES YESQLSLILV DIEAKFYDPS NEDIQFRYIF LKMVFWTARV NLYQCFITLD 420
421 SGILEDEETI IGNLGESCIQ CVRLLISQIT ILEKRGWLLV ALLEIIHALM LAAFCRDKGF 480
481 EVPSDLGDIT LYVQERMVDI VTFDDGMAVR FGYVLRFINS MLHPNEPPMQ DAEPETTEDP 540
541 SKLFADIFDF TSNYFIPSAL LDQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
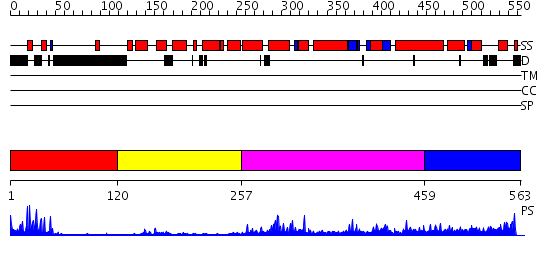
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..119] | 6.30103 | PPR1 | |
| 2 | View Details | [120..256] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [257..458] | 5.446997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [459..563] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)