













| General Information: |
|
| Name(s) found: |
SPAC4G8.03c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 780 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA Golgi apparatus [IDA |
| Biological Process: |
nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay
[ISS |
| Molecular Function: |
mRNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVNRDAYNEL NLNKKSQETN RKPSPLSSYT SISRELDYAN QSPFSSNSFS PTELKARTSA 60
61 TFDVRSASTS PINASDFHKY SHDPFQRLFK AKGNASFSKT KSFPSSVTYS PSEETFPLTS 120
121 GMNKSVHEYP FTLSESAISS SHKSSIPERR NFDSSVSVSN PLLHWNNVDT LLRDGSLENV 180
181 NNSRQDQFLP YKTFSSTISN SDFLHRESSF SSLIDEESKL ASLRNLNIND RPPLPVLKNS 240
241 ERNLLHRQLL SNHPFFSQNN VSLSTNSKNY STDFTKIQSD SSLLQNRQQN HRIETDQLSH 300
301 FPDHLDPSRI PSPYQPSSLQ PLESRKLHSK VDVHSKKLNA LSQLNPILRS ENVLQNDNHH 360
361 SSLSMDNDPT NVSTKNRNNQ TVGEHPYVDD NKKKKKGPAK PKEKATLGKT VNSFFGSHST 420
421 SNYSKVPLSA KLTGEKSDDL SNLLKNKGKK KSQDNQIPHL VGFLGHLSTI CKDQYGCRYL 480
481 QKLLDENPKV NASLFFPEIR QSVVQLMIDP FGNYMCQKLF VYASREQKLS MLNGIGEGIV 540
541 DICSNLYGTR SMQNIIDKLT SNEQISLLLK IIIPSLTTLA CDNNGTHVLQ KCIAKFPPEK 600
601 LEPLFLSMEE NLITLATNRH GCCILQRCLD RTNGDIQERL VNSIIKSCLL LVQNAYGNYL 660
661 VQHVLELNIQ PYTERIIEKF FGNICKLSLQ KFSSNAIEQC IRTASPSTRE QMLQEFLSFP 720
721 NIEQLLDDCY ANYVMQRFLN VADESQKFLI LRSISHVIPK IQNTRHGRHI LAKLTSSTSS 780
781 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
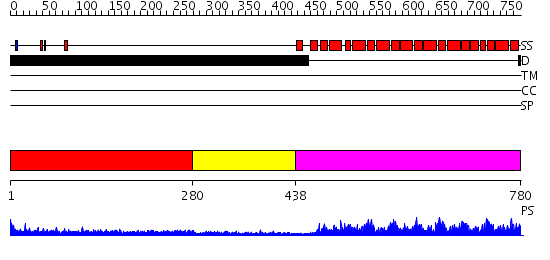
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..279] | 7.447995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [280..437] | 1.436995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [438..780] | 133.0 | Pumilio 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)