













| General Information: |
|
| Name(s) found: |
pol4 /
SPAC2F7.06c
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 506 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
DNA replication
[IEA]
base-excision repair, gap-filling [TAS double-strand break repair via nonhomologous end joining [TAS [NOT]mating type switching [IMP |
| Molecular Function: |
nucleotide binding
[TAS magnesium ion binding [IEA] [NOT]3'-5' exonuclease activity [IDA DNA binding [IEA] DNA-directed DNA polymerase activity [IDA 5'-deoxyribose-5-phosphate lyase activity [IDA DNA-directed RNA polymerase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKILASSTNY VLHNRLSNSQ YEDARSKIVN FGGEFTNDAA KADYIFVNYS QINRVRRELR 60
61 TIGTPLETCV SCKLIVKIDW LNEPKESLTP GNPYVIWHRK PEMKVGSPYT PSTRPASHTE 120
121 APNDFENHET PNTENNNEVK SIDNVDQEGS VYPTTKEYPY VLEIPRYACQ RKTPLKCVNQ 180
181 AFVNALSVLK TCREVNGESV RTRAYGMAIA TIKAFPLPID SAEQLEKMPG CGPKIVHLWK 240
241 EFASTGTLKE AEEFQKDPAS KILLLFYNIF GVGASHAAEW YQKGWRTIEQ VRKHKDSFTK 300
301 QIKVGLEFYE DFCKTVTIEE ATEIYETIVS RMPDGIKIQS CLVGGFRRGK PVGADVDMVL 360
361 SPSHTHSTKH LVDVLLRILD EEFQFRLISV QEHSCGGKKG YVMLAVILSN SSKINRRVDI 420
421 IVVPPAYIGS AVLGWSGGIF FLRDLKLYAN SHLGLSYDSF EIINLKTGKD ICPDEFNEWK 480
481 DPVEAEKDIF RYFSLEYIEP KFRNTG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
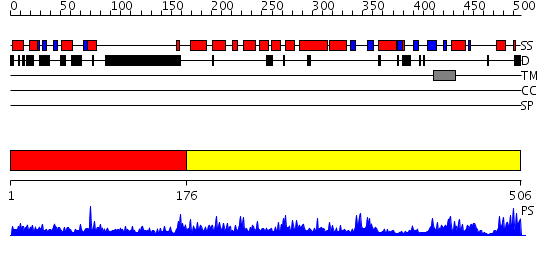
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..175] | 16.30103 | No description for 2dunA was found. | |
| 2 | View Details | [176..506] | 113.0 | Terminal deoxynucleotidyl transferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.48 |
Source: Reynolds et al. (2008)