













| General Information: |
|
| Name(s) found: |
trp2 /
SPAC19A8.15
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 697 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [ISS cytosol [IDA |
| Biological Process: |
tryptophan biosynthetic process
[ISS |
| Molecular Function: |
tryptophan synthase activity
[ISS pyridoxal phosphate binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEQIKKTFL KAKEENKNVL VTFVTCGFPN VDETIKIMQG LQNGGAGIIE LGIPFSDAVA 60
61 DGPTICKGNE IALKNNITLE KVFETVKLAR DAGVTIPIIL MGYYNPIFSY GDAKTIQKAK 120
121 EVGANGFIIV DLPPEEAVGF REECKKQGVS FVPLVAPSTT DRRMELLASV ADSFIYVVSR 180
181 MGSTGSSATG VINTALPQLC QRVRKFAGDT PLAVGFGVNT SEHFHQVGSV SDGVVVGSKI 240
241 IDLILKAEPG TAATVVEEYC KYLTKEDPSA PVPKQFQSGG SVATAPPAAV VEPINEMYLP 300
301 QKYGMFGGMY VPEALTQCLV ELESVFYKAL HDEKFWEEFR SYYEYMGRPS ALDYAKRLTE 360
361 YCGGAHIWLK REDLNHGGSH KINNCIGQIL LAKRLGKNRI IAETGAGQHG VATAICAAKF 420
421 GMKCTIYMGA EDCRRQALNV FRIRLLGAEV VPVTSGTQTL RDAVNEALKA WVEQIDTTHY 480
481 LIGSAIGPHP FPTIVKTFQS VIGEETKAQM QEKRKKLPDA VVACVGGGSN SIGMFSPFKA 540
541 DKSVMMLGCE AGGDGVDTPK HSATLTMGKV GVFHGVRTYV LQREDGQIQD THSISAGLDY 600
601 PGVGPELSEL KYTNRAEFIA VTDAQCLEGF RALCHLEGII PALESSHAVY GGMELAKKLP 660
661 KDKDIVITIS GRGDKDVQSV AEQLPILGPK IGWDLRF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
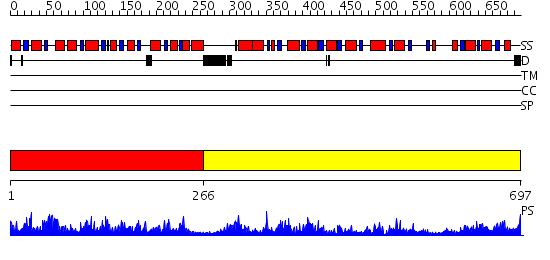
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..265] | 79.522879 | No description for 2ekcA was found. | |
| 2 | View Details | [266..697] | 122.0 | X-ray crystal structure of the Tryptophan Synthase b2 Subunit from Hyperthermophile, Pyrococcus furiosus |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)