













| General Information: |
|
| Name(s) found: |
trp1 /
SPBC1539.09c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 759 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA anthranilate synthase complex [ISS] |
| Biological Process: |
tryptophan biosynthetic process
[ISS]
glutamine metabolic process [IEA] |
| Molecular Function: |
indole-3-glycerol-phosphate synthase activity
[ISS]
phosphoribosylanthranilate isomerase activity [IEA] anthranilate synthase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEKVDVGES VKGDASENAV KEVAERPIVM IDNYDSFTWN VVQYLSNLEK RYPIMVFRND 60
61 EITVDELEKL NPLKLVLSPG PGHPARDGGI CNEAISRFAG KIPILGVCMG LQCIFETMGG 120
121 KVDSAGEIIH GKVSKINHDG LGFYQGIPQN ISVTRYHSLA GKISSLPDCL DVTSWTENGV 180
181 IMGARHKKYA IEGVQYHPES ILSEYGKEYI QNFLNLTAGT WEENGIVMPT KNNAFNAAMR 240
241 ENSNSVSSTK IRKQESILEK IHAQRLIDIA ESKRKPGLSV GDLQTYLNLN IAPPCINFYE 300
301 RLKQSKPALM AEVKRASPSK GDIKLDANAA IQALTYAQVG ASVISVLTEP KWFKGSLNDL 360
361 FVARKAVEHV ANRPAILRKD FIIDPYQIME ARLNGADSVL LIVAMLSREQ LESLYKFSKS 420
421 LGMEPLVEVN CAEEMKTAIE LGAKVIGVNN RNLHSFEVDL STTSKLAEMV PDDVILAALS 480
481 GISSPADVAH YSSQGVSAVL VGESLMRASD PAAFARELLN LSSSEISNGK KTSTPLVKVC 540
541 GTRSLLAAKT IVESGGDLIG LIFVEKSKRK VDLSVAKEIS HFVHTTNRKH ISPKKAVTGQ 600
601 SWFDHQYENL ASSPHPLLVG VFQNQPLEYI RSIIAEVNLD IVQLHGQEPF EWIHMLDRPV 660
661 IKVFPLNSSE ISRPNYHIVP LIDAYVGGES GGLGKKVDWE AASFIPVSYV LAGGLTPKNV 720
721 QDAISVSRPA VVDVSSGVET DGKQDLEKIK AFINAVKEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
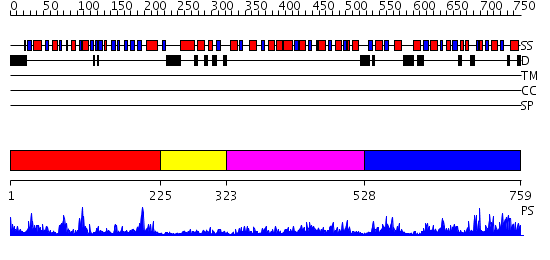
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..224] | 39.69897 | Anthranilate synthase GAT subunit, TrpG | |
| 2 | View Details | [225..322] | 3.30103 | No description for 2dplA was found. | |
| 3 | View Details | [323..527] | 25.154902 | REFINED STRUCTURE OF SPINACH GLYCOLATE OXIDASE AT 2 ANGSTROMS RESOLUTION | |
| 4 | View Details | [528..759] | 101.0 | THREE-DIMENSIONAL STRUCTURE OF THE BIFUNCTIONAL ENZYME PHOSPHORIBOSYLANTHRANILATE ISOMERASE: INDOLEGLYCEROLPHOSPHATE SYNTHASE FROM ESCHERICHIA COLI REFINED AT 2.0 ANGSTROMS RESOLUTION |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)