













| General Information: |
|
| Name(s) found: |
taf1 /
SPAC7D4.04
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 926 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA pre-autophagosomal structure [ISS] extrinsic to membrane [ISS] |
| Biological Process: |
protein localization to pre-autophagosomal structure
[ISS]
induction of conjugation upon nitrogen starvation [IMP autophagy [ISS] peroxisome degradation [ISS] telomere maintenance [ISS protein targeting to vacuole [ISS CVT pathway [ISS] cellular response to nitrogen starvation [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFRLNDSFS GKSWTIDNVQ WTPQNLVAWI MELNIGLSDE AKLLLPNGMS LNETVLNSES 60
61 DVVIYVLDQN LLTFTYDGDK IPSIPSLKGQ PISNVLTGII ADSSSDQWKD KISKCLSLLS 120
121 DILESSSKIH NQLSVCHDEY STSIVSPEVA MNYLQRRQSG MKDLLFVFYE RLDRVSVSDL 180
181 LHDFLALPTG SLPITPLKIL STNWTKLDSW LNSISARYAE AQKRVQQCIG AANSIIVSPF 240
241 KEVPSIKEGD DAYYHLRSVA QDAANILQEI LKIESTSTTS QIKPKCNYET EITDCMEKLQ 300
301 SNLSELKSSR KSSLISLKSF WLSFYKVSLR YDALYEYLRQ IAEELDRSKF VLSQSRNIFS 360
361 LYIDILMEAL RRTEWQESYN VSHDSSLPLD QEKELSLRKS WLSHFSNLLF NHGQLKYLPV 420
421 LTRDEISNYL LNIQAHPNYQ TFHQMLSNRL SEYLGFPGSP REAVSNTVQA NNSLHEKLAM 480
481 YQNRCNNLEA MLSHQNGFNY NINNDGLSPN APHPPINEQS NSSQPFYRVS PSIVPLNVIR 540
541 KLTNRKTSFT DSHILRLQDE VNQLRNELDL VNKRNEDLLI ELQGKEEKIQ YLETENEEVL 600
601 QKYENLQEEL SSTRKLLTKN EAAVAEQQSN EEMHSNEPNI LNLYSVFEGK VDSLQELYNA 660
661 FKLQLTSLKQ KGEYATLAKD AEAVQHIIEE RDYALAEKAD LLKLSENRKE QCKILTQKLY 720
721 TIVFRCNELQ SVLRECVTQP GFDSDNERDG HASIPDRQIE FNSKDLQYLY WMDGEDADRN 780
781 FQEFLNRMSS LDFDSFHNFV VSVLTQAHNH ELRWKREFQS NRDKALKAIL DSQSKVSLRN 840
841 FKQGSLVLFL PTRRTAGNKK VWAAFNVNAP HYYLNTQPHL KLESRDWMLG RVTSIEDRTA 900
901 DDSTDKWLRL PSGTIWHLVE AIDERF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
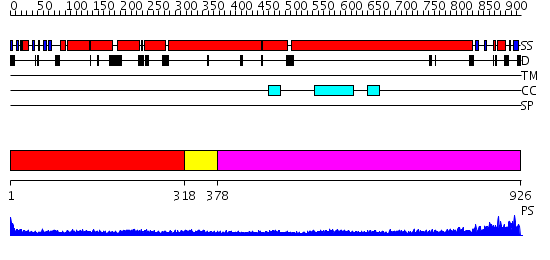
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..317] | 15.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [318..377] | 9.09691 | Tropomyosin | |
| 3 | View Details | [378..926] | 1.23 | COLICIN IA |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)