













| General Information: |
|
| Name(s) found: |
SPAC23A1.12c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [ISS cytosol [IDA phenylalanine-tRNA ligase complex [IC |
| Biological Process: |
phenylalanyl-tRNA aminoacylation
[ISS |
| Molecular Function: |
magnesium ion binding
[IEA]
ATP binding [IEA] RNA binding [IEA] phenylalanine-tRNA ligase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTISCDKEE LYKALGREYT TQEFDELCFQ FGIELDEDTT NDPERSPSER PSLKIDIPAN 60
61 RYDMLCLEGI AQALNVFNRR MATPQYKLLP STTSLTISPE TSEIRPYAAA AILRGVKLDP 120
121 IRYQSFIALQ DKLHANLCRN RTLVAIGTHD FSVMEGPFTY EALKPEEINF VPLNQTQEIN 180
181 GSNLLEFYKD SKHLSRYLHI IANSPRYPVI LDAKRRVCSL PPIINSEFSK ISVDTRDIFI 240
241 DVTATDKTKL EIVVNMMTTM FSCYCEEPFT IEPVNIISEH NGCTRVTPNL NPTCFKADID 300
301 YLNEACGLSL PEDEICHLLT RMMLTAKPNP NDSKTLLVYV PPLRADILHQ CDIMEDLGIA 360
361 YGYDNLKHTY PAHSVTFGKP FEVNRLADII RNEVAYAGWS EVMPFILCSH DENYAWLRKT 420
421 DDSKAVQLAN PKTLEFQVVR SSLLPGILKT VRENKNHALP IKIFEVSDVA FCDYSRERMT 480
481 RNERHLCAIF AGLNSGFEQI HGLLDRVMLM LNTKRIMNPK DSDAVGYWIE AEDDSTFFPG 540
541 RCAAVYYRKD FGTAGIRVGV FGVLHPLVLE KFELTSAASA VEIDLTLWV |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Unpublished Data |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
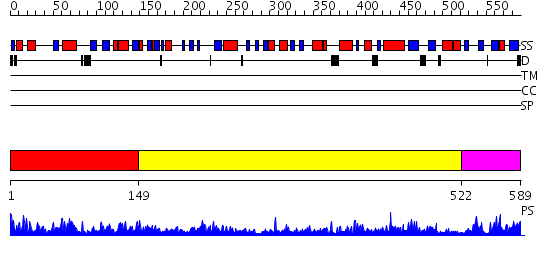
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | 72.0 | No description for 2cxiA was found. | |
| 2 | View Details | [149..521] | 5.045757 | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with an analogue of threonyl adenylate | |
| 3 | View Details | [522..589] | 60.39794 | Phenyl-tRNA synthetase (PheRS) alpha subunit, PheS |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)