













| General Information: |
|
| Name(s) found: |
swi6 /
SPAC664.01c
[Sanger Pombe]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 328 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mating-type region heterochromatin
[IDA nucleus [IDA nuclear telomeric heterochromatin [IDA spindle pole body [IDA centromeric heterochromatin [TAS |
| Biological Process: |
sister chromatid cohesion
[IGI chromatin remodeling [RCA chromatin silencing at centromere [IMP chromatin assembly or disassembly [IEA] chromatin silencing at silent mating-type cassette [IMP donor selection [IMP chromatin silencing at telomere [IMP mating type switching [IMP cellular protein localization [IGI |
| Molecular Function: |
protein binding
[IPI chromatin binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKKGGVRSYR RSSTSKRSVI DDDSEPELPS MTKEAIASHK ADSGSSDNEV ESDHESKSSS 60
61 KKLKENAKEE EGGEEEEEDE YVVEKVLKHR MARKGGGYEY LLKWEGYDDP SDNTWSSEAD 120
121 CSGCKQLIEA YWNEHGGRPE PSKRKRTARP KKPEAKEPSP KSRKTDEDKH DKDSNEKIED 180
181 VNEKTIKFAD KSQEEFNENG PPSGQPNGHI ESDNESKSPS QKESNESEDI QIAETPSNVT 240
241 PKKKPSPEVP KLPDNRELTV KQVENYDSWE DLVSSIDTIE RKDDGTLEIY LTWKNGAISH 300
301 HPSTITNKKC PQKMLQFYES HLTFRENE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
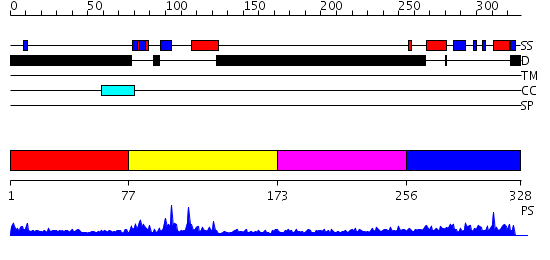
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [77..172] | 9.30103 | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | |
| 3 | View Details | [173..255] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [256..328] | 21.154902 | Heterochromatin protein 1, HP1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)