













| General Information: |
|
| Name(s) found: |
skb1 /
SPBC16H5.11c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 645 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cell tip [IDA cell cortex [IDA |
| Biological Process: |
regulation of response to osmotic stress
[IMP unidimensional cell growth [IGI establishment or maintenance of cell polarity [IGI negative regulation of mitotic cell cycle [IGI peptidyl-arginine N-methylation [IC cellular protein localization [IGI |
| Molecular Function: |
protein binding
[IPI protein-arginine N-methyltransferase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLRDGRIPQ YSIYPLPVPP MNSKTPTLGI VCSEGTISLS LEEGFEFVGV PLSGEGLKLR 60
61 VEALAPSERL QEFLDDEVAY HPEENVHKVV GLSSAWLELD SEDTLIADRS EEVLLKEASY 120
121 ASYCGLSSII LNGPTSPMNV MRYARAVSSA LNSTMNLKFL VQLAIESGHE DYFETWKMWD 180
181 TIRSACGYHP RLKVALELPP ACSPPIELVN RWYAEPIEMI TMSCMAFVPN PNGYPVLGRK 240
241 LRAIYALYLR LNPRILLWDN DAPEKIGDSP DYSIYMKHLF DSQPPAPLVE DLADSYKDYL 300
301 QVPLQPLSYN LENITYEIFE RDPVKYAQYE QAIFSALMDR DESSVTRIAV VGAGRGPLVD 360
361 CALRAAISSS RTVDMIALEK NPNAFSMLLM RNRQDWAGKV TLVFGDMRTW NPDYKIDILV 420
421 SELLGSMGDN ELSPECLDGV QHVLDEETGI CIPSSYISYV TPIMSPKLWS EARNMNDPNA 480
481 FERQYVVLMN SFDFLAADDE FRFQSLWSFH HPNKDSEVYT KNLHNKRFAS VRFQASSPGI 540
541 LHGFAGYFEA TLYKDISLSI MPATMEAKSP DMFSWFPIYM PIKKPMYVPE NSQLEFHMWR 600
601 LTDGMRVWFE WCANAYLVLR NGSQIKLSST EVHNISGKAF SCNMY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
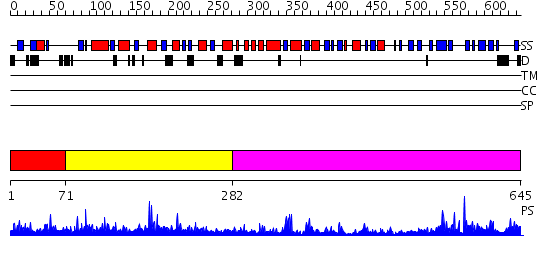
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [71..281] | 1.314976 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [282..645] | 73.0 | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)