













| General Information: |
|
| Name(s) found: |
SPAC1F12.07
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 389 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[ISS cytosol [IDA |
| Biological Process: |
L-serine biosynthetic process
[ISS |
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
O-phospho-L-serine:2-oxoglutarate aminotransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSREEVVNF AAGPAAMITS VVEEFGKDFV NFQGLGMGVA EISHRSKQGS GIVTSAESNF 60
61 RKLYNIPENF HILFMQGGGT EQFAACLYNV YAHHALKNGN AKSLVANYII TGAWSKKAYA 120
121 EAERLGFPCH VAVDMKELAG KYGSLPEDKD LKFTPDGETS LVYYCDNETV HGVEFNEPPT 180
181 NIPKGAIRVC DVSSNFISRK IDFTKHDIIF AGAQKNAGPA GITVVFVRDS VLARPTPAEL 240
241 HKLNIPVSPT VSDYKIMADN HSLYNTLPVA TLHAINLGLE YMLEHGGLVA LEASSIEKSK 300
301 LLYDTLDKHD LYISVVEPAA RSRMNVTFRI EPQELESEFL AEAEKHHLVQ LKGYRSVGGI 360
361 RASLYNAISV EQTRRLIDLL ESFAKAHSN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
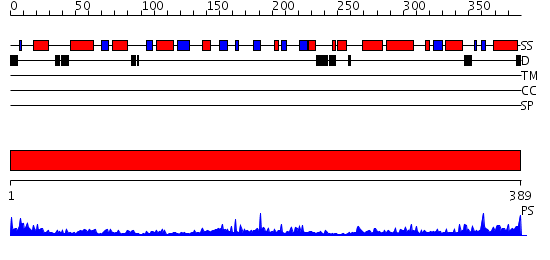
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..389] | 52.0 | Crystal structure of phosphoserine aminotransferase from Bacillus alcalophilus |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)