













| General Information: |
|
| Name(s) found: |
Tf2-8 /
SPAC19D5.09c
[Sanger Pombe]
Tf2-7 / SPAC13D1.01c [Sanger Pombe] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1333 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS cellular_component [ND] |
| Biological Process: |
biological_process
[ND]
proteolysis [IEA] DNA integration [IEA] RNA-dependent DNA replication [IEA] |
| Molecular Function: |
DNA binding
[IEA]
RNA-directed DNA polymerase activity [IEA][RCA RNA binding [IEA] aspartic-type endopeptidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYANYRYMK ARAKRWRPEN LDGIQTSDEH LINLFAKILS KHVPEIGKFD PNKDVESYIS 60
61 KLDQHFTEYP SLFPNEHTKR QYTLNHLEEL EQQFAERMFS ENGSLTWQEL LRQTGKVQGS 120
121 NKGDRLTKTF EGFRNQLDKV QFIRKLMSKA NVDDFHTRLF ILWMLPYSLR KLKERNYWKS 180
181 EISEIYDFLE DKRTASYGKT HKRFQPQNKN LGKESLSKKN NTTNSRNLRK TNVSRIEYSS 240
241 NKFLNHTRKR YEMVLQAELP DFKCSIPCLI DTGAQANIIT EETVRAHKLP TRPWSKSVIY 300
301 GGVYPNKINR KTIKLNISLN GISIKTEFLV VKKFSHPAAI SFTTLYDNNI EISSSKHTLS 360
361 QMNKVSNIVK EPELPDIYKE FKDITAETNT EKLPKPIKGL EFEVELTQEN YRLPIRNYPL 420
421 PPGKMQAMND EINQGLKSGI IRESKAINAC PVMFVPKKEG TLRMVVDYKP LNKYVKPNIY 480
481 PLPLIEQLLA KIQGSTIFTK LDLKSAYHLI RVRKGDEHKL AFRCPRGVFE YLVMPYGISI 540
541 APAHFQYFIN TILGEVKESH VVCYMDNILI HSKSESEHVK HVKDVLQKLK NANLIINQAK 600
601 CEFHQSQVKF IGYHISEKGF TPCQENIDKV LQWKQPKNRK ELRQFLGSVN YLRKFIPKTS 660
661 QLTHPLNNLL KKDVRWKWTP TQTQAIENIK QCLVSPPVLR HFDFSKKILL ETDASDVAVG 720
721 AVLSQKHDDD KYYPVGYYSA KMSKAQLNYS VSDKEMLAII KSLKHWRHYL ESTIEPFKIL 780
781 TDHRNLIGRI TNESEPENKR LARWQLFLQD FNFEINYRPG SANHIADALS RIVDETEPIP 840
841 KDSEDNSINF VNQISITDDF KNQVVTEYTN DTKLLNLLNN EDKRVEENIQ LKDGLLINSK 900
901 DQILLPNDTQ LTRTIIKKYH EEGKLIHPGI ELLTNIILRR FTWKGIRKQI QEYVQNCHTC 960
961 QINKSRNHKP YGPLQPIPPS ERPWESLSMD FITALPESSG YNALFVVVDR FSKMAILVPC1020
1021 TKSITAEQTA RMFDQRVIAY FGNPKEIIAD NDHIFTSQTW KDFAHKYNFV MKFSLPYRPQ1080
1081 TDGQTERTNQ TVEKLLRCVC STHPNTWVDH ISLVQQSYNN AIHSATQMTP FEIVHRYSPA1140
1141 LSPLELPSFS DKTDENSQET IQVFQTVKEH LNTNNIKMKK YFDMKIQEIE EFQPGDLVMV1200
1201 KRTKTGFLHK SNKLAPSFAG PFYVLQKSGP NNYELDLPDS IKHMFSSTFH VSHLEKYRHN1260
1261 SELNYATIDE SDIGTILHIL EHKNREQVLY LNVKYISNLN PSTIMSGWTT LATALQADKA1320
1321 IVNDYIKNNN LNI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
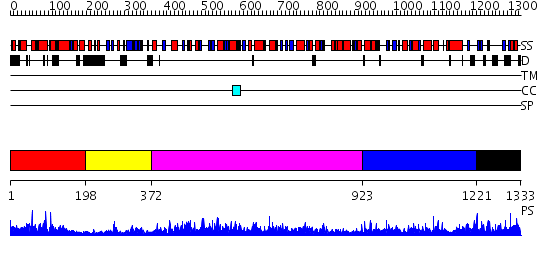
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [198..371] | 25.69897 | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | |
| 3 | View Details | [372..922] | 162.0 | HIV RNase H (Domain of reverse transcriptase); HIV-1 reverse transcriptase | |
| 4 | View Details | [923..1220] | 70.221849 | N-terminal Zn binding domain of HIV integrase; Retroviral integrase, catalytic domain | |
| 5 | View Details | [1221..1333] | 1.31 | Polycomb chromodomain complexed with the histone H3 tail containing trimethyllysine 27. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)