













| General Information: |
|
| Name(s) found: |
rng3 /
SPCC613.04c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 746 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA actomyosin contractile ring [IDA cytosol [IDA actin filament [IDA |
| Biological Process: |
cytokinesis, actomyosin contractile ring assembly
[IMP 'de novo' cotranslational protein folding [IPI cellular protein localization [IGI |
| Molecular Function: |
protein binding
[IPI myosin binding [ISS mRNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTHELSSTPQ IDLLNDILKN SVESNVFSDY QKKQIVTLIL KSKISTAIVL LSPKSTAASE 60
61 WNYLCNLQDL HECVLCVDTI LPANLQTIAK RIFSLVLLPP LNDWCKQLRD AFLRFVSQPS 120
121 ICPTDFPLKL FFLSTVGIEL LIVNEKIIPQ KTQKYLLYEL FSSPSSITAN EIARLCQEAN 180
181 NRNYLLQSLT SATDARRAFL NNPNYRLLSA IIFQDGPSNL AFVLAKDCVL LARQPETIPV 240
241 SFERMSSLLL MCLPSHPDFI SPEFCHEWTE LAERNGLQEE WLNILNTACN FKECRAIIHK 300
301 ECSEFIKDNH TSRVAILISM KLAFQYQLSQ VIPTLKLLLQ SKVYDSVLLE ALRQSSTLGP 360
361 VKQLIADDSC LLNNLSKLLL DTNISPLDAS SIATIIYNMC KFKITKSEHE RELNQLRNMA 420
421 EASKTIDYKE DETAPTERRI QKILEYDILS KLFSAAKHYN SLNGLLAMIL VHMANYKLAR 480
481 RKLVQIGALK FLTRQCFIQT QDSNAAFALA KILISVAPHS IFTKAFPSNR AIHPMSKLLS 540
541 TNSADTEYPI LLGKFEVLLA LTNLASHDEE SRQAIVQECW RELDELIIET NPLIQRATTE 600
601 LINNLSLSPY CLIKFIGDKD SDFENTRLHI VLALSDTEDT PTRLAACGIL VQITSVDEGC 660
661 KKILSLQNDF NYIVRMLTDQ DEGIQHRGLV CICNIVYSKD QEIFNKFIKT PKAVETLRTY 720
721 ITKQAALKEL QHEALVMIDS RLQGSK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
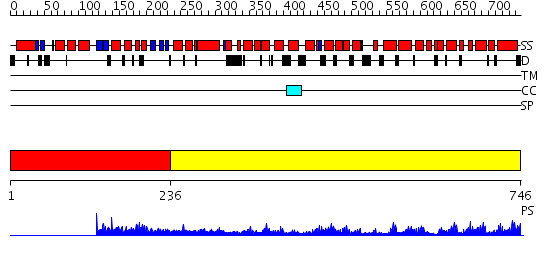
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..235] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [236..746] | 71.0 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)