













| General Information: |
|
| Name(s) found: |
rec10 /
SPAC25G10.04c
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 791 amino acids |
Gene Ontology: |
|
| Cellular Component: |
linear element
[IDA nucleus [IDA |
| Biological Process: |
linear element assembly
[IMP reciprocal meiotic recombination [IMP meiotic sister chromatid segregation [IMP meiotic sister chromatid cohesion [IMP meiotic DNA double-strand break formation [IGI cellular protein localization [IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDIFSEFLNC LSSDGTLNES SIYKTYQILE SLNPKDVDTK ENYIKLSNTF STLGSGVGFQ 60
61 DNLLIEMFKI LTVLFFKTRS TDLGDLLIES FTSLEIEKLM RVKKTIGSIV LTKGIQELQE 120
121 IELPKVGFNC MTYDESIFQG ISLERLILQL MSIFIAKCEE NKLWLVNNRK DLDLRVIGQR 180
181 LLHRDFACRF LSGLFISRFS VSGDTDESKH QNGIRLQLFI DFSKFETRLT MQILKADRIF 240
241 STCVANTVHA YEGLFSSGSN IDSTIATLVL EPRDLIVYRK GFALLQIPWT SVTTIDKIKK 300
301 VKSLKIITEV SSLDEFVFQC KDGDKFDELF STSEEIMNKL LPAIIVSPKL TLRNRSIIQI 360
361 KEGESKLPNT SKQASQNLPH LDDELAYQRF EDQVIDKSVC DDECTNTENT PSSNIPADVK 420
421 DSLSADDYAY DTKRKTQIED LEEDQNKSKI ASKDGTNLKE INSVPEFSDE NVINQTGPAK 480
481 KTPVQRRKDG KFAKSTKRKK QKSLKPDTEN QESSVKNKKA KSNVNLQYSP KTPICKINDE 540
541 TLKPPTIANI AGHKQMNHLT SENIETPVPV PNGNWYNGVK HETATDIFTT CHDGNNSLKS 600
601 SVWKELLKEK HWKQESKPQL TGNSRQIDLS TFVKQANTPN ITSLLDGTCS SPPNNECFND 660
661 KEPDSSSSTL ISDRQELEYR NPNAETVKLE EIPYNKFFKT VEKNEAYNPS SKSATIDGLQ 720
721 RYTSMIGNQI YEGILQNEKE LRSKLEAYHI NCNKVIKEFS KRQTARYKII EKELAQIEVN 780
781 LVSQIDSLMF K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
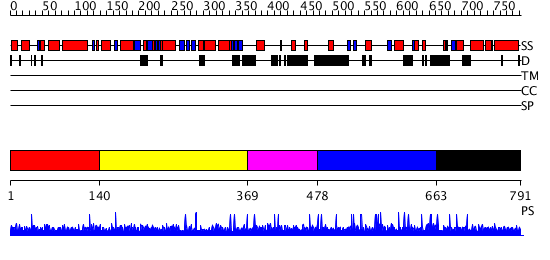
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..139] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [140..368] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [369..477] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [478..662] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [663..791] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)