













| General Information: |
|
| Name(s) found: |
SPCC1259.09c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 456 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial pyruvate dehydrogenase complex
[ISS mitochondrion [IDA |
| Biological Process: |
acetyl-CoA biosynthetic process from pyruvate
[ISS |
| Molecular Function: |
lipoic acid binding
[NAS acyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKHYIHQCV KASSCKHSLS VKQRYFHCSA LNGVASMFRM PALSPTMEEG NITKWHFKEG 60
61 DSFKSGDILL EVETDKATMD VEVQDNGILA KVLIEKGSNI PVGKNIAIVA DAEDNLKDLE 120
121 LPKDEASSEE QSFSSSKEEV KPIVQDRETK SNVEHKSTSQ ANDAVNKSFL PSVSYLIHQY 180
181 KIENPWSIPA TGPHGRLLKG DVLAHVGKID KGVPSSLQKF VRNLEVLDFS GVEPKKPSYT 240
241 EDSVPVRKST MIETLKTVPM KDEYITFERS ISLEKLKSLL AKRKEKDAFG IDEVVSAMIG 300
301 DALSISELSK VEPNSIESAY DLLLGAPIVK LKSSVSNFQP KLFPKNNNTK DLHKYIRDSM 360
361 IGLPSNLSDY VKLNFSSVTK SSSNNVEFLD DVYDMLLGSN SDSIDSRINR HEEPSTTEDL 420
421 TLTLSIKDNI SKSRAIKFVD CLKSNFENPE FVLSRW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
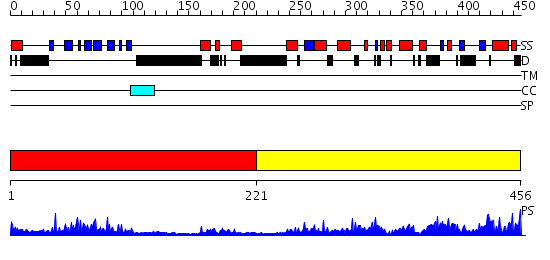
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..220] | 23.522879 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 2 | View Details | [221..456] | 63.30103 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)