













| General Information: |
|
| Name(s) found: |
SPBC776.15c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 452 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial oxoglutarate dehydrogenase complex
[IC mitochondrion [IDA mitochondrial nucleoid [ISS |
| Biological Process: |
tricarboxylic acid cycle
[ISS 2-oxoglutarate metabolic process [ISS |
| Molecular Function: |
dihydrolipoyllysine-residue succinyltransferase activity
[ISS lipoic acid binding [IEA] oxidoreductase activity [RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSYGNGFRM MAKCLLSLRS GYSVTAPVSK SMANVLWARY ASTRIKTPPF PESITEGTLA 60
61 QWLKQPGEYV NKDEEIASVE TDKIDAPVTA PDAGVLKEQL VKEGDTITID QDIAVIDTSA 120
121 APPEGGSAGP KKDEVKTADA DAAKDLSTPQ DSSKPIEEKP MPDLGAEQKE SAPSSTKPAP 180
181 DAKEPEFSSP KPKPAKSEPV KQSKPKATET ARPSSFSRNE DRVKMNRMRL RIAERLKESQ 240
241 NRAASLTTFN ECDMSAVVAL RKKYKDEILK ETGVKIGFMS FFSKACTQAM KQIPAINGSI 300
301 EGEGKGDTLV YRDFCDLSIA VATPKGLVTP VIRNAESMSL LEIESAIATL GSKARAGKLA 360
361 IEDMASGTFT ISNGGIFGSL YGTPIINLPQ TAVLGLHAIK ERPVVINGQV VPRPMMYLAL 420
421 TYDHRMVDGR EAVTFLRLVK EYIEDPAKML LV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
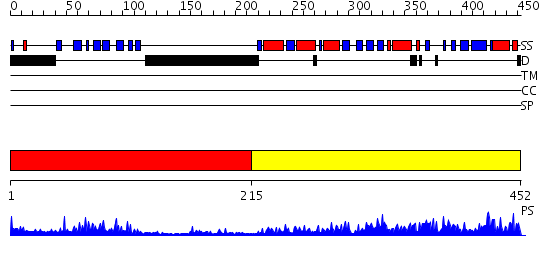
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..214] | 22.39794 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 2 | View Details | [215..452] | 87.30103 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)