













| General Information: |
|
| Name(s) found: |
mkh1 /
SPAC1F3.02c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1116 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell septum
[IDA cell division site [IDA cell tip [IDA cytoplasm [IDA cytosol [IDA |
| Biological Process: |
positive regulation of calcium-mediated signaling
[IMP MAPKKK cascade [IDA cellular response to heat [IMP endoplasmic reticulum unfolded protein response [ISS] cell morphogenesis [NAS cellular cell wall organization [IMP response to osmotic stress [IMP cellular sodium ion homeostasis [NAS cytokinesis [NAS protein amino acid phosphorylation [ISS |
| Molecular Function: |
ATP binding
[ISS protein binding [IPI MAP kinase kinase kinase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAADIGSQSS GSLEERFEQS LHLQNVDKQD WSLNSVLQFL KLYKFNKEWE DVFIKSRIEM 60
61 DLFINLADQS KAEEFAFKNK LSKESAIQLS SCIRKTLLAP SSTRVPSKNS SYETLTYSAK 120
121 DSSDDVFTET NSGFRSSNQN SSLKSFQSVP DSNVNVFGGF GGSVVDNNEL LSTGKNSHQT 180
181 TSLNLEGSPI NLHAYKGTVT SIINDDSRNI NKKTLSKQPV SEHKEKQTSF LRRFRVPGFS 240
241 RDKDKTKDCP SSNSNPFHLA SSNVKTLDAS LDQGEWVPRI HRLESQIGLI SKKKSFVLAT 300
301 MDDMKFTVVD ITNVQNATQL RKLIAKSMYL DISIDQFDLF LTEVGGAQYI EILDDRKLDI 360
361 ARLYSDEFGT IKFFVKPSQN EESGMDSDTY LSFGTKSSST YKADDDSIYH RKEDFKKQPS 420
421 YPVLTSDFEI TDAGPNLSLS GHQPDNKYYK GFSSAPNLAV VPELPSRRFR GFEKIRGAKG 480
481 EMATKILDAT EAQSEKNKFT VCRPHKKVTL KMPLNSGSSA PQSPSSNTSA SVLTRNFVAH 540
541 RDPPPPPTET SSLRRKNTLT RRPSIRHARS SPYIDTGHNE ASKFSHTSFD PKASSKSSNS 600
601 LKESVEALSE IPFEDAPALD ESDLSGDPFW AIQPKQSSSQ VPKENHHNIQ SKLSINTEAA 660
661 TDLKANELSS PKTPEYCRGD DRSISLSPLS YRLRKSKHIR ESPPSSKVIN SGNWEVRPSA 720
721 DDLYEDVDRF FPRYDLDKVL VVDQSRMVSS PSKVSIRPKM KSVRLLAREA SEARKEIRHN 780
781 ARRNKSGNLL RRSSTKLWGS RIVELKPDTT ITSGSVVSQN ATFKWMKGEL IGNGTYGKVF 840
841 LAMNINTGEL IAVKQVEIPQ TINGRHDQLR KDIVDSINAE ISMIADLDHL NIVQYLGFEK 900
901 TETDISIFLE YVSGGSIGRC LRNYGPFEEQ LVRFVSRQVL YGLSYLHSKG IIHRDLKADN 960
961 LLIDFDGVCK ISDFGISKHS DNVYDNDANL SMQGSIFWMA PEVIHNDHQG YSAKVDVWSL1020
1021 GCVVLEMLAG RRPWSTDEAI QAMFKLGTEK KAPPIPSELV SQVSPEAIQF LNACFTVNAD1080
1081 VRPTAEELLN HPFMKCDEEF NFKDTNLYDM LCKRKS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
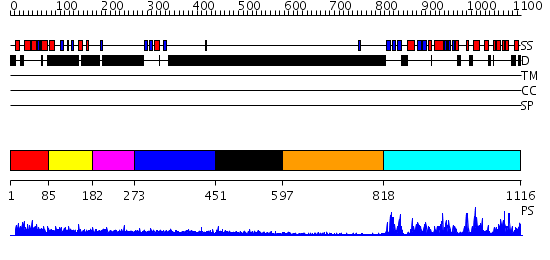
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 1.17 | Sterile alpha motif (SAM) domain of mouse bifunctional apoptosis regulator | |
| 2 | View Details | [85..181] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [182..272] | 3.151996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [273..450] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [451..596] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [597..817] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [818..1116] | 64.30103 | No description for 2clqA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)