













| General Information: |
|
| Name(s) found: |
mal3 /
SPAC18G6.15
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 308 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA microtubule cytoskeleton [IDA spindle midzone [IDA cytoplasm [TAS kinetochore microtubule [IDA cell cortex of cell tip [IDA cytosol [IDA cytoplasmic microtubule [IDA astral microtubule [IDA |
| Biological Process: |
regulation of filamentous growth
[IMP spindle assembly [IMP regulation of mitosis [IGI cell morphogenesis [IMP attachment of spindle microtubules to kinetochore [IGI regulation of catalytic activity [IC microtubule polymerization [IMP microtubule-based movement [IMP microtubule depolymerization [ISS cellular protein localization [IGI |
| Molecular Function: |
protein binding
[IPI microtubule plus-end binding [IDA ATPase activator activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSESRQELLA WINQVTSLGL TRIEDCGKGY AMIQIFDSIY QDIPLKKVNF ECNNEYQYIN 60
61 NWKVLQQVFL KKGIDKVVDP ERLSRCKMQD NLEFVQWAKR FWDQYYPGGD YDALARRGNR 120
121 GPANTRVMNS SAGATGPSRR RQVSSGSSTP SMTKSSANNN NVSSTANTAA VLRAKQAQQQ 180
181 ITSLETQLYE VNETMFGLER ERDFYFNKLR EIEILVQTHL TTSPMSMENM LERIQAILYS 240
241 TEDGFELPPD QPADLTTALT DHDTNNVAEE AQMTDLKDSE TQRVPSAPDF VHARLQSLEV 300
301 DDDENITF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
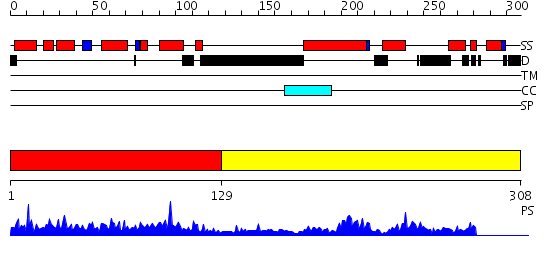
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 63.69897 | Solution structure of the CH domain of human microtubule-associated protein RP/EB family member 3 | |
| 2 | View Details | [129..308] | 25.0 | Crystal structure of the EB1 C-terminal domain complexed with the CAP-Gly domain of p150Glued |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)