













| General Information: |
|
| Name(s) found: |
tif32 /
SPBC17D11.05
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 932 amino acids |
Gene Ontology: |
|
| Cellular Component: |
eukaryotic 43S preinitiation complex
[IDA cytoplasm [IDA eukaryotic translation initiation factor 3 complex [ISS |
| Biological Process: |
positive regulation of translational initiation
[IC regulation of translation in response to nitrogen starvation [IPI translational initiation [ISS |
| Molecular Function: |
translation initiation factor activity
[ISS protein binding [IPI arylformamidase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPPQGKPEN VLRLADELIA LDQHSSALQS LHETIVLKRS RNAQGFSLEP IMMRFIELCV 60
61 HLRKGKIAKE GLYTYKNAVQ NTSVTAIENV VKHFIELANK RVQEAQEKAD KISVEYVDDL 120
121 EATETPESIM MSLVSGDLSK SRTDRALVTP WLKFLWDAYR TVLDILRNNA RLEVMYQLIA 180
181 NSAFQFCLKY QRKTEFRRLC ELLRSHLGNA SKFSNAPHSI NLNDAETMQR HLDMRFSQLN 240
241 VAVELELWQE AFRSIEDIHS LLTFSKRAPA AVMLGNYYRK LIKIFLVCDN YLLHAAAWNR 300
301 YFTFTNVQKP ATANFVILSA LSIPIIDANK LSGPSIEAED AKSKNARLAL LLNLSKTPTR 360
361 ETLIKDAISR GVLSFCDQAI RDLYQILEVE FHPLSICKKL QPIIKRLAES NDTAQYIRPL 420
421 QQVILTRLFQ QLSQVYDSIS LKYVMDLATF EEPYDFNPGQ IEKFIMNGNK KGAFSIRLNH 480
481 IENSISFSSD LFSNPIKSSD SVSLQSTPSE LITSQLTRIA KSLSSVLMRF DTDFCLLRKQ 540
541 QAEAAYERAQ AGVEQERKAV IAQRSLLELR RGQADTLATQ REAELAAQRA LKQKQESEAE 600
601 SLRVQEEINK RNAERIRREK EAIRINEAKK LAEELKAKGG LEVNAEDLEH LDADKLRAMQ 660
661 IEQVEKQNKS MNERLRVIGK RIDHLERAYR REAIPLWEED AKQQAEHDRE IFYEREKQRK 720
721 EVQERKHEQA IKDKKAFAQF ASYIHAYKQN IDDERDKAYQ EAYAKAKNVI DAERERQRKE 780
781 IFEQKLAEAI REAEEEAARA AEEEANRELH EQEEAQKRAI EERTRAAREA KEREQREMAE 840
841 KLERQRRIQQ ERDEEISRKL AEKAAARRAN IGASSPSPGA WRRGGASAGG VSRDSPRYSR 900
901 GGYSRGSVPP RETLAPSKGA YVPPSRRNQQ QQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
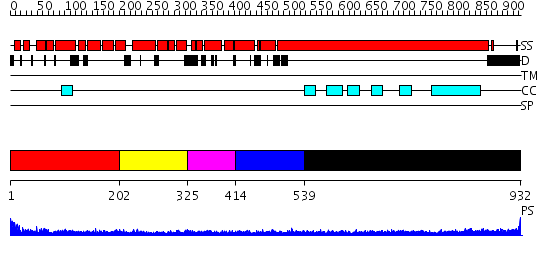
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..201] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [202..324] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [325..413] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [414..538] | 2.1 | Solution structure of the PCI domain | |
| 5 | View Details | [539..932] | 14.69897 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)