













| General Information: |
|
| Name(s) found: |
hsp90 /
SPAC926.04c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 704 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[ISS mitochondrion [ISS cytosol [IDA |
| Biological Process: |
conjugation with cellular fusion
[IMP cellular response to stress [ISS negative regulation of transcription by glucose [IDA response to nutrient [IMP protein folding [IMP cAMP-mediated signaling [IMP |
| Molecular Function: |
ATP binding
[IEA]
unfolded protein binding [IEA] ATPase activity, coupled [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNTETFKFE AEISQLMSLI INTVYSNKEI FLRELISNAS DALDKIRYQS LSDPHALDAE 60
61 KDLFIRITPD KENKILSIRD TGIGMTKNDL INNLGVIAKS GTKQFMEAAA SGADISMIGQ 120
121 FGVGFYSAYL VADKVQVVSK HNDDEQYIWE SSAGGSFTVT LDTDGPRLLR GTEIRLFMKE 180
181 DQLQYLEEKT IKDTVKKHSE FISYPIQLVV TREVEKEVPE EEETEEVKNE EDDKAPKIEE 240
241 VDDESEKKEK KTKKVKETTT ETEELNKTKP IWTRNPSEVT KEEYASFYKS LTNDWEDHLA 300
301 VKHFSVEGQL EFRAILFVPR RAPMDLFEAK RKKNNIKLYV RRVFITDDCE ELIPEWLGFI 360
361 KGVVDSEDLP LNLSREMLQQ NKIMKVIRKN LVRRCLDMFN EIAEDKENFK TFYDAFSKNL 420
421 KLGIHEDAAN RPALAKLLRY NSLNSPDDLI SLEDYITKMP EHQKNIYFIT GESKQAVENS 480
481 PFLEIFRAKK FDVLFMVDPI DEYAVTQLKE FEGKKLVNIT KDGLELEETD EEKAAREKLE 540
541 KEYEEFAKQL KTILGDKVEK VVVSNKIVGS PCLLTTGQYG WSANMERIMK AQALRDTSMS 600
601 AYMSSRKTFE INPKSPIIAE LKKKVEENGA EDRSVKDLAT ILYETALLSS GFTLDDPSAY 660
661 AQRINRLISL GLSIDEEEEA PIEEISTESV AAENNAESKM EEVD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
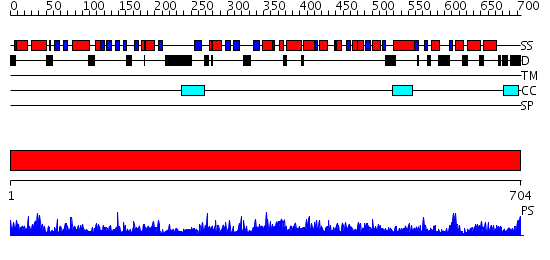
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..704] | 1000.0 | No description for 2cg9A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)