













| General Information: |
|
| Name(s) found: |
SPAC13G6.06c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1017 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA glycine cleavage complex [ISS |
| Biological Process: |
glycine decarboxylation via glycine cleavage system
[ISS |
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
glycine dehydrogenase (decarboxylating) activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFRACSKLQY HGVNTSLSRH LFLAKRNLSI SSACLEAKNS QKFPALDTFE PRHIGPSKTD 60
61 QQYQLESLGY KDFDSFLKDV IPDSVRTPES QLMAFGSVNP NEKNPPVNYS ESEFTTLANN 120
121 VANQNKLIKS FIGMGYYNVK LPAAIQRNVL ENPEWYTQYT PYQAEISQGR LESMMNYQTM 180
181 IADLTGLSIS NASLLDEGTA AGEAMVMLMA NDKKKRKTFL VDKNIYPNTL SVLRTRASGF 240
241 GIKIELDNIT PELITKSAKH VFGIFVQYPA ADGSIFDYGH LAATARSFNM HVVAATDLLA 300
301 LTILKSPGEW GADVAVGSTQ RFGLPMGYGG PHAGFFACSE EFKRKIPGRL IGLSKDRLEN 360
361 PAYRLALQTR EQHIRREKAT SNICTAQALL ANMSAFYAIY HGPNGLQEIA NRIYASTSFL 420
421 KSALESSGYK IVNKSHFFDT LTIEVESADK VLAKALDHGY NLRKVDDSHV GLSLDETVCD 480
481 KDIQALFSIF NINKSVDQYY MEIATSEPNG NSASTVDNLS ICSLPENFRR TTLYLQHPVF 540
541 NRYHSETELM RYIHHLQSKD LSLAHAMTPL GSCTMKLNAV TEMMPITNPL FANIHPYVPE 600
601 EQAKGYRHVI EDLQLMLTTI TGFDAACFQP NSGAAGEYTG LSVIRAYQRS IGQGHRNICL 660
661 IPVSAHGTNP ASAAMAGFTV IPVKCLNNGY LDMQDLKEKA SKHADKLAAF MVTYPSTFGI 720
721 FEPDVKEALE VIHEHGGQVY FDGANMNAMV GLCKAGDIGA DVCHLNLHKT FCIPHGGGGP 780
781 GVGPICVKKH LADFLPSHPV VSCGGKNGIT SVSSSPFGSA GILPISWAYM RMMGLAGLRD 840
841 ASKAALLNAN YMAKRLSSHY KLVYTNKNNL CAHEFILDAR EFKATAGVDA TDIAKRLQDY 900
901 SFHAPTLSWP IANTLMIEPT ESESMYEMDR FCDALISIRQ EIREIEEGLQ PKDNNLLVNA 960
961 PHPQKDIASE KWDRPYTRER AVYPVPLLKE RKFWPSVARL DDAYGDKNLF CTCSPVV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
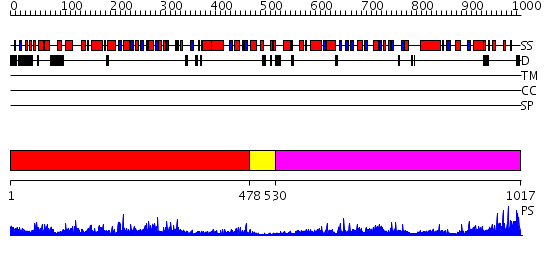
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..477] | 61.154902 | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in apo form | |
| 2 | View Details | [478..529] | 16.69897 | No description for 2qmaA was found. | |
| 3 | View Details | [530..1017] | 58.09691 | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in apo form |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)