













| General Information: |
|
| Name(s) found: |
fus1 /
SPAC20G4.02c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1372 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cytoplasm [IDA mating projection [IDA cytosol [IDA |
| Biological Process: |
plasma membrane fusion during cytogamy
[IMP cellular response to stress [IEP actin cytoskeleton organization [ISS actin filament bundle assembly [NAS |
| Molecular Function: |
actin binding
[ISS SH3 domain binding [IEA] Rho GTPase binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMTASFKGIR ISKPIVKEIS SFGSLTNIDL HTDPYEKVEA KALSRSRLKN QSVKKTDLRI 60
61 TNDYSSLFVS IENKKNTIPD IHCNNLSKRP VGFTDSYVPG EALLCPDDPN LVNELFDDVL 120
121 KRLREEKSDS GEQFHWDANV TLSQENKWSF ITGYLSRNDK FLKISSNDSE LNVKASVISY 180
181 IEKLSCTPLK LKYVESLAVA LRTESVTWVR YFLLMGGHIV LAKILQAIHE KKHLKSPDIT 240
241 LEIAILKSMR CIIGQKIGTD FYLSNKYPID SVVHCLLSTK LLVRKLAAEL LTFLCYSEKP 300
301 NGINAIMKCM KNFANLKVES MNVFDLWLSQ WKRTLLEKVV TEEKKVLEEA ALHDKITIEY 360
361 IISQMLLINA FLENIPSKTA LLQFKESLRI GNFKSILLIL KKINDEGVLK QLEKCVKLVS 420
421 LDTANEKHFL KHTPNSAAHQ SLLNTNMFND ANFEFMVKEH IKNFLKLLKE HNNPVRIIKL 480
481 LDCLVLTLQA DKGNGNSNND IDCFYKELKI GNEQGNNAPG YSSVTSGYGT TNSKKVVASY 540
541 DNTDDNEYSV SKSELYATGD TNNTTNQGYE NSERKYVESS KYETDLKNIF DCLCLLFPSE 600
601 FDPQSSFSAE KIFSKLKHLL TRTRDSYISE DTKILRLLNR SSSKLQDRDQ EYLISKNNVN 660
661 GNGNRDWVQP VNTTSSVSAF ASPRIKPKLL FQPNQDEKLK LCKLDLAKAN SITLKSEPSS 720
721 AVSTHSFEVH LSMPGTQIKS ANLAEKMETS KHKVFNPKRI DVVSDLPLDY RKSYYGRFSI 780
781 TDTKRFSKIE NMRIKEVIDG NPFKAPPPAP LPPPAPPLPT AMSSLQKFEK NDSQIFRKTI 840
841 IIPENISIDD IFKFCSGSES EVYASKIPGE LCNPSKRLKQ LHWKRLEVPF EKTLWNIVVA 900
901 DPYLLTLKLT AEGIFKQLED CYPLREVTVS NKKVKEYTGF MPVDLQQMVS IRLHRFNSLT 960
961 PIEIAKKFFH CDHDIMELVD FFNDRKFFRQ EGLKKQLKPY MSSRNEEVME VSEKLLELSR1020
1021 FDQIYTLIVV DIDTYYEKRM AALKIKSFLA NNFRDFRRQI RKLHCASLEL KSSLHFKYFL1080
1081 NLVLHIGNFM NDAPRRAKGY RLESLLRASM IKNDKTGLTL LHTIEKIVRT HFPQLEAFLV1140
1141 DLKAIPEISR FNLEQLEQDC NDICERMKNV EKDFSNEGIF SNHKALHPDD HICEVMVPWI1200
1201 PSGKSMVDEL NSDITELKTT LKTTLLMYGE NPDEPTSSAR FFNNLNEIIL EYKKASTVNQ1260
1261 KMEKEEELAF LRLQALKASV KSNNAENSGS SKNEEVSATM ENLISQLQKG LCDDSLSNKT1320
1321 DCTESSKQTI ETLMNYENDK QTLEGSNKKR QTVVLKAECM LKQLENNNEL KR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
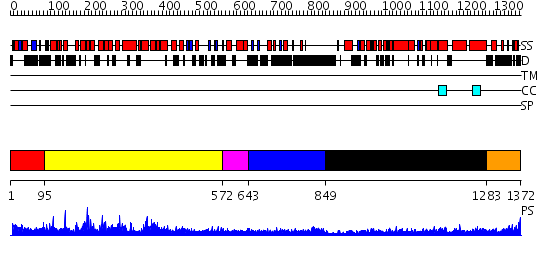
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [95..571] | 77.522879 | Crystal structure of mDIA1 GBD-FH3 in complex with RhoC-GMPPNP | |
| 3 | View Details | [572..642] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [643..848] | 3.307994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [849..1282] | 38.39794 | Bni1p Formin Homology 2 Domain complexed with ATP-actin | |
| 6 | View Details | [1283..1372] | 5.09691 | Interferon-induced guanylate-binding protein 1 (GBP1), C-terminal domain; Interferon-induced guanylate-binding protein 1 (GBP1), N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)