













| General Information: |
|
| Name(s) found: |
cut8 /
SPAC17C9.13c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 262 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA nucleus [IDA nuclear membrane [TAS |
| Biological Process: |
proteasomal ubiquitin-dependent protein catabolic process
[IMP mitotic spindle organization [IMP anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process [IGI chromosome segregation [IMP proteasome localization [IMP response to DNA damage stimulus [IGI mitotic metaphase/anaphase transition [IMP |
| Molecular Function: |
polyubiquitin binding
[IDA protein anchor [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METLSYSQIK KRKADFDEDI SKRARQLPVG EQLPLSRLLQ YSDKQQLFTI LLQCVEKHPD 60
61 LARDIRGILP APSMDTCVET LRKLLINLND SFPYGGDKRG DYAFNRIREK YMAVLHALND 120
121 MVPCYLPPYS TCFEKNITFL DAATNVVHEL PEFHNPNHNV YKSQAYYELT GAWLVVLRQL 180
181 EDRPVVPLLP LEELEEHNKT SQNRMEEALN YLKQLQKNEP LVHERSHTFQ QTNPQNNFHR 240
241 HTNSMNIGND NGMGWHSMHQ YI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
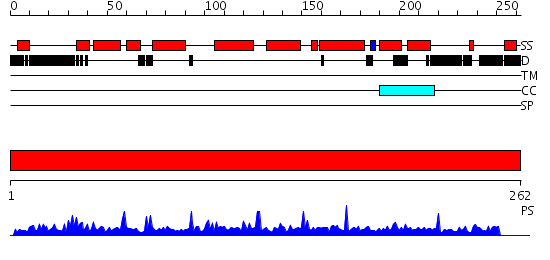
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..262] | 100.49485 | No description for PF08559.1 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)