













| General Information: |
|
| Name(s) found: |
csx2 /
SPBC17G9.08c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 870 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA plasma membrane [IDA |
| Biological Process: |
small GTPase mediated signal transduction
[IC response to arsenic [IMP regulation of GTPase activity [ISS regulation of ARF GTPase activity [IEA] response to cadmium ion [IMP intracellular protein transport [RCA |
| Molecular Function: |
zinc ion binding
[IEA]
ARF GTPase activator activity [IEA] GTPase activator activity [ISS phospholipid binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTGHVDPVHF ERLKIQSVTF GSANDNASLQ VHNVSKSPEY SKYSYSRKNN EPIEFIQLRE 60
61 EEGPIVKVEE EDGLTLRFQL EFHAQEGKRD VTNLTCVYGT SPQQLDRLIT REFSSDANFQ 120
121 NNHQVFLIGN IEFHSNENSK KIEWTWSNQS LHTLYFRNGG SPIYLCFAEY DKRLNCLVPL 180
181 KTFQFYVTES DQDSTPATPF PMPMHLAEET ATSPEISDAP PLSGNVDPLP INSPPLTNPV 240
241 ARDIDQTEPE DGPLFRATIL NYERTTHDMR MVLKKLIKRI EHVAHSHGLL YMSYKELMSA 300
301 FERVATINPP AFKPFLDHYY AAAAQSFDSF NIDRARLLRN FLIEPLRKIY DTDIKNVSTK 360
361 KKDFEETSRD YYTSLSRYLS KSEKETSSDK TKESKFAAKK RDFELSRFDY YSYMQDINGG 420
421 RKGQEVLSVL TSFAANDYNL IHSSLTDIDA LRPSIIQLQD IVTEANKEFQ LLRAEREERR 480
481 RYIETSSREL EDKDAEIAAQ AYKAKVDQDT SAKQGLLLAF SKSTSDLQVV GKSGWHKYWV 540
541 VLDHGKICEY ANWKQSLELH TEPIDLLMAT VRPAQSVSRK FCFEVITPQT KRTYQATSKA 600
601 EMHSWIEAIQ YSISESIVQK GKGTSMNSEE TSVKHGPTST IGKALQRVAS VTSPSRHNSD 660
661 SKEKKQTKSP SLVKTLKEMH SSDQSCADCN TTARVEWCAI NFPVVLCIDC SGIHRSLGTH 720
721 ITKIRSLTLD KFNPETVDLL YATGNSFVNE IYEGGITDWK IKNFENQERR VQFVKDKYLY 780
781 KRFIKSSFSH DPNTGLLESI EHSNLKEAVL CLALGADVNY QRAVVKALLK NNYLIAELLT 840
841 LNGASLNVDE VTMETLSARA QAFVMNRATP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
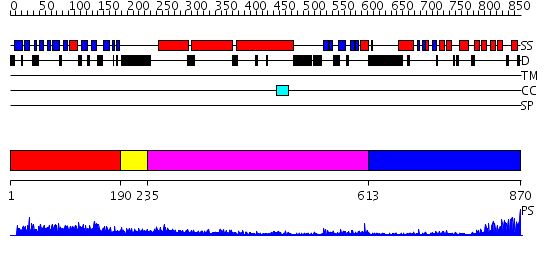
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..189] | 16.69897 | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | |
| 2 | View Details | [190..234] | 12.09691 | Heavy meromyosin subfragment | |
| 3 | View Details | [235..612] | 19.69897 | No description for 2elbA was found. | |
| 4 | View Details | [613..870] | 15.0 | Pyk2-associated protein beta; Pyk2-associated protein beta ARF-GAP domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)