













| General Information: |
|
| Name(s) found: |
csx1 /
SPAC17A2.09c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 632 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA cytosol [IDA |
| Biological Process: |
regulation of mRNA stability involved in response to stress
[IMP |
| Molecular Function: |
RNA binding
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIDCLYRRS SLFDTSFVPL HSSIPATSKM SASNSDVNAP ISPVVDEGKS ELVSPTLERL 60
61 VAPFNCSPSS TPLQDVAGVG SKMSDTLWMG DLEPWMDATF IQQLWASLNE PVNVKVMRSK 120
121 ASSSETLISY CFVQFSSSAA AERALMKYNN TMIPGAHCTF KLNWATGGGI QHNNFVSRDP 180
181 EFSIFVGDLL PTTEDSDLFM TFRSIYPSCT SAKIIVDPVT GLSRKYGFVR FSSEKEQQHA 240
241 LMHMQGYLCQ GRPLRISVAS PKSRASIAAD SALGIVPTST SNRQPNQDLC SMDPLNTTVF 300
301 VGGLASNLSE KDLQVCFQPF GRILNIKIPF GKGCGFVQYS EKSAAEKAIN TMQGALVGTS 360
361 HIRLAWGHNT LPVSALSQSQ SQVSDEGFDR TLSANQIFGM NQSVIGANSG SSNSSGSSLK 420
421 SAPVSPRTAA AQSLLPNSVV SSINGMNSVN FSTISPPPLS RSASISPTLS GSGSGLTPLS 480
481 SHFPSAATGL VGGQVYPQSS VLQSSKINGS AKVQPSVKLP EWLQPFSGNN HNSFATQDLL 540
541 TRVSSLKLVD DEQPASLNGS AFQARASRPW NLGRERQSSL IDLRHELEQN ENGLEKSGFG 600
601 LNLRGRLPPR SYSTFNCTGQ YLQPSLRLSR DS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
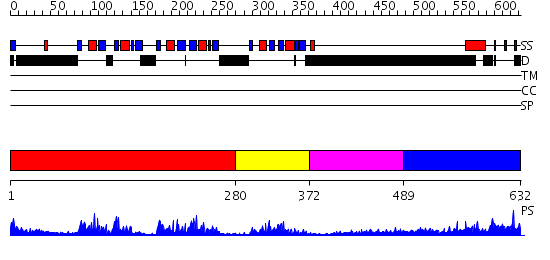
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..279] | 49.221849 | Crystal Structure of UP1 Complexed With d(TTAGGGTTAG(6-MI)G); A Human Telomeric Repeat Containing 6-methyl-8-(2-deoxy-beta-ribofuranosyl)isoxanthopteridine (6-MI) | |
| 2 | View Details | [280..371] | 17.0 | Polypyrimidine tract-binding protein | |
| 3 | View Details | [372..488] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [489..632] | 21.0 | poly(A) binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)