













| General Information: |
|
| Name(s) found: |
clr3 /
SPBC800.03
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 687 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mating-type region heterochromatin
[IDA nucleus [IDA nuclear chromatin [IDA telomeric heterochromatin [IDA NuRD complex [IDA histone deacetylase complex [NAS rDNA heterochromatin [IDA centromeric heterochromatin [IDA nucleolar chromatin [IDA |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IMP regulation of histone methylation [IMP chromatin silencing at centromere [IMP chromatin silencing at rDNA [IMP nucleosome positioning [IMP chromatin silencing at silent mating-type cassette [IMP histone deacetylation [IMP chromatin silencing at telomere [IMP regulation of transcription from RNA polymerase II promoter [IEP |
| Molecular Function: |
histone deacetylase activity
[IDA histone deacetylase activity (H3-K14 specific) [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLASNSDGAS TSVKPSDDAV NTVTPWSILL TNNKPMSGSE NTLNNESHEM SQILKKSGLC 60
61 YDPRMRFHAT LSEVDDHPED PRRVLRVFEA IKKAGYVSNV PSPSDVFLRI PAREATLEEL 120
121 LQVHSQEMYD RVTNTEKMSH EDLANLEKIS DSLYYNNESA FCARLACGSA IETCTAVVTG 180
181 QVKNAFAVVR PPGHHAEPHK PGGFCLFNNV SVTARSMLQR FPDKIKRVLI VDWDIHHGNG 240
241 TQMAFYDDPN VLYVSLHRYE NGRFYPGTNY GCAENCGEGP GLGRTVNIPW SCAGMGDGDY 300
301 IYAFQRVVMP VAYEFDPDLV IVSCGFDAAA GDHIGQFLLT PAAYAHMTQM LMGLADGKVF 360
361 ISLEGGYNLD SISTSALAVA QSLLGIPPGR LHTTYACPQA VATINHVTKI QSQYWRCMRP 420
421 KHFDANPKDA HVDRLHDVIR TYQAKKLFED WKITNMPILR DSVSNVFNNQ VLCSSNFFQK 480
481 DNLLVIVHES PRVLGNGTSE TNVLNLNDSL LVDPVSLYVE WAMQQDWGLI DINIPEVVTD 540
541 GENAPVDILS EVKELCLYVW DNYVELSISK NIFFIGGGKA VHGLVNLASS RNVSDRVKCM 600
601 VNFLGTEPLV GLKTASEEDL PTWYYRHSLV FVSSSNECWK KAKRAKRRYG RLMQSEHTET 660
661 SDMMEQHYRA VTQYLLHLLQ KARPTSQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
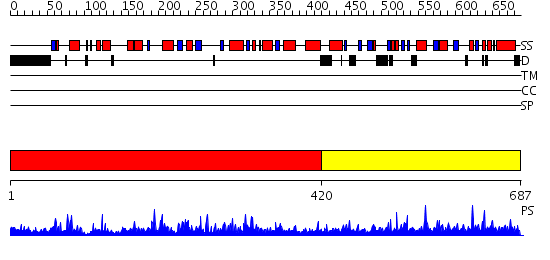
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..419] | 103.0 | No description for 2nvrA was found. | |
| 2 | View Details | [420..687] | 1.12 | No description for 2r8bA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)