













| General Information: |
|
| Name(s) found: |
cdr2 /
SPAC57A10.02
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 775 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA medial cortex [IDA medial cortical node [IDA spindle pole body [IDA |
| Biological Process: |
signal transduction
[RCA regulation of cell size [IGI regulation of cytokinesis [IMP protein amino acid phosphorylation [IC cellular protein localization [IMP negative regulation of G2/M transition of mitotic cell cycle [IGI regulation of G2/M transition of mitotic cell cycle [IMP |
| Molecular Function: |
ATP binding
[ISS protein binding [IPI protein serine/threonine kinase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTISEVGPW ELGLSLGSGG PNSSRLAKHR ETGQLAVVKP IVGWSELTSS QQARIEGELV 60
61 LLRLIEHPNV LQLIDVISAQ EQLFVVVEYM PGGELFDCML RKGSFTEQDT AKFLWQILCG 120
121 LEYCHKLHIC HRDLKPENLY LDAHGSIKIG EFGMASIQQP GKLLQTSCGS PHYASPEIIM 180
181 GRSYDGCASD IWSCGIIFFA LLTGKLPFDD DNIRSLLLKV CQGQFEMPSN ISPQAQHLLY 240
241 RMLDVDSSTR ITMEQIREHP FLSCFVHPNI SIPIISAPIQ PIDPLIVQHL SLVFRCSDDP 300
301 MPLYEKLASQ SPLVEKTLYT LLSRHLHPPS SAAVDRNRAV VDDLLGTAAS NGQQMDEEEI 360
361 EQAINIPTLA PYPISYAAES VPRPATSASP FLTPVTTSGT FNYSFNATNP QSILQRPATT 420
421 SSAVPQLPKS VTPGLAYPHD SSMLSSNYRP PSALSPRNFN VSINDPEVQL SRRATSLDMS 480
481 NDFRMNENDP SIVGNLAASN FPTGMGPPRK RVTSRMSEHT GNRVVSFPRG SAFNPRVTRF 540
541 NVGNEQFSNN IDNNNYNQPY ANATMNNSRR LRTPSGERSM RADLSQSPAS YDSLNVPKHR 600
601 RRQSLFSPSS TKKKLSGSPF QPKRSFLRRL FSSEPSCKCV YASLVASELE HEILEVLRRW 660
661 QLLGIGIADI IYDSVSASIS ARIKRQNSLN LKPVRFRISV LAEFFGSQAV FVLESGSSTT 720
721 FDHLATEFQL IFEDKGFLDN LELSYFQASA SRPVSRMSVS SSPFAVFRQR QSVQS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
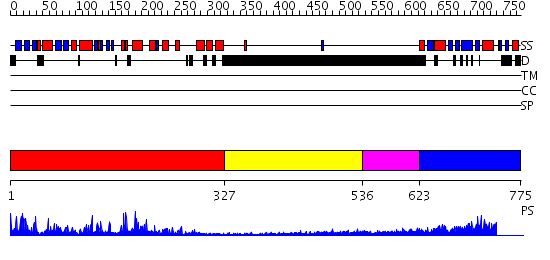
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..326] | 82.09691 | Catalytic and ubiqutin-associated domains of MARK2/PAR-1: Wild type | |
| 2 | View Details | [327..535] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [536..622] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [623..775] | 36.0 | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)