













| General Information: |
|
| Name(s) found: |
atf1 /
SPBC29B5.01
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 566 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [NAS |
| Biological Process: |
regulation of histone methylation
[IMP regulation of cAMP-mediated signaling [IMP regulation of histone acetylation [IMP regulation of transcription, DNA-dependent [TAS cellular response to starvation [IMP response to hydrogen peroxide [IGI negative regulation of reciprocal meiotic recombination [IMP cellular response to nitrogen starvation [IMP activation of reciprocal meiotic recombination [IMP [NOT]response to arsenic [IGI regulation of chromatin disassembly [IMP regulation of chromatin assembly [IMP G1 phase of mitotic cell cycle [IMP mRNA export from nucleus in response to heat stress [IMP gene-specific transcription from RNA polymerase II promoter [RCA positive regulation of transcription, meiotic [IDA regulation of reciprocal meiotic recombination [IMP |
| Molecular Function: |
DNA binding
[TAS transcription factor activity [IDA RNA binding [IDA protein binding [IPI protein homodimerization activity [IPI protein heterodimerization activity [IPI specific RNA polymerase II transcription factor activity [RCA recombination hotspot binding [IDA sequence-specific DNA binding [IDA promoter binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPSPVNTST EPASVAAVSN GNATASSTQV PENNQSDSFA PPSNNSQQNQ QSSTIAPNGG 60
61 AGSVANANPA DQSDGVTPSF VGSLKLDYEP NPFEHSFGST ASVGQGNPSL NRNPSLSNIP 120
121 SGVPPAFART LLPPVSSIAS PDILSGAPGI ASPLGYPAWS AFTRGTMHNP LSPAIYDATL 180
181 RPDYLNNPSD ASAAARFSSG TGFTPGVNEP FRSLLTPTGA GFPAPSPGTA NLLGFHTFDS 240
241 QFPDQYRFTP RDGKPPVVNG TNGDQSDYFG ANAAVHGLCL LSQVPDQQQK LQQPISSEND 300
301 QAASTTANNL LKQTQQQTFP DSIRPSFTQN TNPQAVTGTM NPQASRTQQQ PMYFMGSQQF 360
361 NGMPSVYGDT VNPADPSLTL RQTTDFSGQN AENGSTNLPQ KTSNSDMPTA NSMPVKLENG 420
421 TDYSTSQEPS SNANNQSSPT SSINGKASSE SANGTSYSKG SSRRNSKNET DEEKRKSFLE 480
481 RNRQAALKCR QRKKQWLSNL QAKVEFYGNE NEILSAQVSA LREEIVSLKT LLIAHKDCPV 540
541 AKSNSAAVAT SVIGSGDLAQ RINLGY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
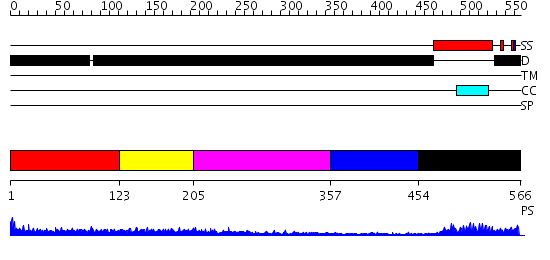
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | 1.158995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [123..204] | 3.166995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [205..356] | 4.171995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [357..453] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [454..566] | 15.69897 | C/ebp beta |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)