













| General Information: |
|
| Name(s) found: |
fma1 /
SPBC3E7.10
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 379 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosolic ribosome [ISS cytosol [IDA nucleolus [IDA |
| Biological Process: |
protein initiator methionine removal
[ISS]
proteolysis [ISS |
| Molecular Function: |
metalloexopeptidase activity
[IEA]
aminopeptidase activity [ISS cobalt ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATEIAKHIC CGIDCNNEAD RLQCPKCLND GVKSYFCGQE CFRNSWNIHK HLHRPPNVEK 60
61 REDGTYNPFP KFHFAGSLKP VYPLSPIRKV PPHIKRPDYA KTGVSRSEQI EGRSFKLKRL 120
121 TPKEQEGMRK VCRLGREVLD AAAAAVRPGT TTDELDSIVH NACIERDCFP STLNYYAFPK 180
181 SVCTSVNEII CHGIPDQRPL EDGDIVNIDV SLYHNGFHGD LNETYYVGDK AKANPDLVCL 240
241 VENTRIALDK AIAAVKPGVL FQEFGNIIEK HTNSITEKQI SVVRTYCGHG INQLFHCSPS 300
301 IPHYSHNKAP GIARPGMTFT IEPMLTLGPA RDITWPDDWT SSTASGRCSA QFEHTLLVTE 360
361 TGCEVLTARL PNSPGGPLK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
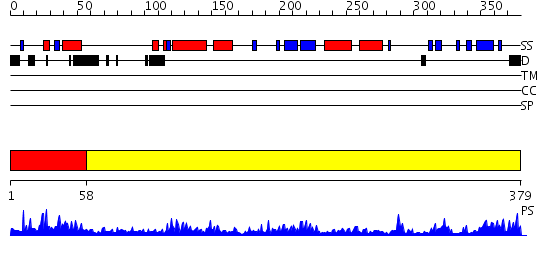
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..57] | 3.107992 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [58..379] | 79.69897 | Crystal structure of Human Methionine Aminopeptidase Type I with a third cobalt in the active site |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)