













| General Information: |
|
| Name(s) found: |
FBpp0309477
[FlyBase]
Ubqn-PA / FBpp0074519 [FlyBase] |
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 547 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
amyloid precursor protein metabolic process
[IMP]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEGGSKRIN VVVKTPKDKK TVEVDEDSGI KDFKILVAQK FEAEPEQLVL IFAGKIMKDT 60
61 DTLQMHNIKD NLTVHLVIKA PTRNNEQPAR QPADVRQTPF GLNQFGGLAG MEALGAGSNT 120
121 FMDLQARMQN ELLNNGDMLR SLMDNPMVQQ MMNNPDTMRQ LITSNPQMHD LMQRNPEISH 180
181 MLNNPDLLRQ TMELARNPSM LQELMRSHDR AMSNLESVPG GYSALQRIYR DIQEPMMNAA 240
241 TESFGRNPFA GLVDGGGSGA GNNPQQGTEN RNPLPNPWGG ANSGTNGTVG GSGAGNPTGD 300
301 LPPNNVLNTP AMRSLLQQMA DNPAMMQNLL NAPYTRSMME SMSQDPDMAA RLLSSSPLMS 360
361 NNPALQEQVR QMMPQFMAQM QNPEVMNMLT NPDAMNAILQ IQQGMEQLRS AAPGLVGTLG 420
421 IPPPPPGAGT GTNPASGDGS GGNSGASTNN VSPSSGLNAG TGTPNLAPGG GPNAQLFNDF 480
481 MMRMLNGMSN NADNTQPPEV RYQSQLEQLN AMGFVNRDAN LQALIATFGD INAAVERLLS 540
541 LNQLSLS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
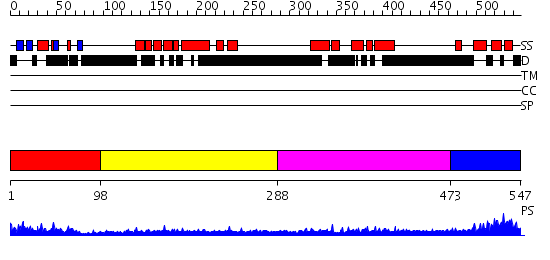
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | 17.0 | Solution Structure of the N-terminal Ubiquitin-like Domain in the Human Ubiquilin 3 (UBQLN3) | |
| 2 | View Details | [98..287] | 0.973 | Crystal structure of human pregnenolone sulfotransferase (SULT2B1a) in the presence of PAP | |
| 3 | View Details | [288..472] | 6.522879 | Sec24 | |
| 4 | View Details | [473..547] | 8.522879 | Solution Structure of RSGI RUH-016, a UBA Domain from mouse cDNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)