













| General Information: |
|
| Name(s) found: |
B-H1-PA /
FBpp0074204
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 544 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][NAS]
|
| Biological Process: |
sensory organ boundary specification
[IMP]
compound eye photoreceptor cell differentiation [IMP] bristle morphogenesis [IMP] negative regulation of transcription [IGI][IMP] eye-antennal disc morphogenesis [IMP] regulation of transcription, DNA-dependent [IEA][NAS] eye pigment granule organization [IMP] regulation of transcription [IEA] leg disc proximal/distal pattern formation [IMP] |
| Molecular Function: |
DNA binding
[NAS]
transcription factor activity [IEA][NAS] specific RNA polymerase II transcription factor activity [NAS] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKDSMSILTQ TPSEPNAAHP QLHHHLSTLQ QQHHQHHLHY GLQPPAVAHS IHSTTTMSSG 60
61 GSTTTASGIG KPNRSRFMIN DILAGSAAAA FYKQQQHHQQ LHHHNNNNNS GSSGGSSPAH 120
121 SNNNNNINGD NCEASNVAGV GVLPSALHHP QPHPPTHPHT HPHALMHPHG KLGHFPPTAG 180
181 GNGLNVAQYA AAMQQHYAAA AAAAAARNNA AAAAAAAAAA AAAGVAAPPV DGGVDGGVGL 240
241 APPAGGDLDD SSDYHEENED CDSGNMDDHS VCSNGGKDDD GNSVKSGSTS DMSGLSKKQR 300
301 KARTAFTDHQ LQTLEKSFER QKYLSVQERQ ELAHKLDLSD CQVKTWYQNR RTKWKRQTAV 360
361 GLELLAEAGN FAAFQRLYGG SPYLGAWPYA AAAGAAHGAT PHTNIDIYYR QAAAAAAMQK 420
421 PLPYNLYAGV PSVGVGVGVG VGPAPFSHLS ASSSLSSLSS YYQSAAAAAS AANPGGPHPV 480
481 APPPSVGGGS PPSGLVKPIP AHSASASPPP RPPSTPSPTL NPGSPPGRSV DSCSQSDDED 540
541 QIQV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
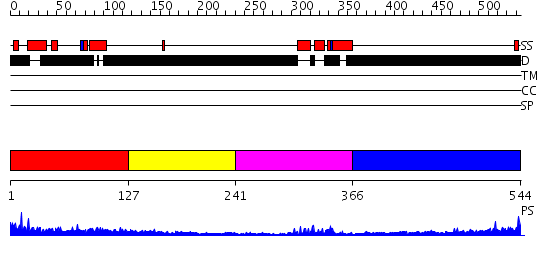
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 1.20599 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [127..240] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [241..365] | 11.30103 | Homeobox protein hox-b1 | |
| 4 | View Details | [366..544] | 1.201987 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.42 |
Source: Reynolds et al. (2008)