













| General Information: |
|
| Name(s) found: |
HDAC6-PB /
FBpp0073827
[FlyBase]
|
| Description(s) found:
Found 34 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1135 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
|
| Biological Process: |
histone deacetylation
[IDA]
regulation of transcription [IDA] |
| Molecular Function: |
zinc ion binding
[IEA]
histone deacetylase activity [IDA][ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRDIWIQSP PIVTRRSAQQ AKIQTRAMAS KSKTGTTGAA TAAGGTGSSS GSISGGFNAA 60
61 DPRKSNKPNA ALLEAKRRAR NNMLKNQGCG SQESVTDIFQ NAVNSKSLVR KPTALIYDES 120
121 MSQHCCLWDK EHYECPERFT RVLERCRELN LTERCLELPS RSATKDEILR LHTEEHFERL 180
181 KETSGIRDDE RMEELSSRYD SIYIHPSTFE LSLLASGSTI ELVDHLVAGK AQNGMAIIRP 240
241 PGHHAMKAEY NGYCFFNNVA LATQHALDVH KLQRILIIDY DVHHGQGTQR FFYNDPRVVY 300
301 FSIHRFEHGS FWPHLHESDY HAIGSGAGTG YNFNVPLNAT GMTNGDYLAI FQQLLLPVAL 360
361 EFQPELIIVS AGYDAALGCP EGEMEVTPAC YPHLLNPLLR LADARVAVVL EGGYCLDSLA 420
421 EGAALTLRSL LGDPCPPLVE TVPLPRAELA QALLSCIAVH RPHWRCLQLQ QTYDCVELQD 480
481 RDKEEDLHTV LRHWIGGPPP MDRYPTRDTA IPLPPEKLTS NAARLQVLRA ETKLSVPSFK 540
541 VCYAYDAQML LHCNLNDTGH PEQPSRIQHI HKMHDDYGLL KQMKQLSPRA ATTDEVCLAH 600
601 TRAHVNTVRR LLGREPKELH DAAGIYNSVY LHPRTFDCAT LAAGLVLQAV DSVLRGESRS 660
661 GICNVRPPGH HAEQDHPHGF CIFNNVAIAA QYAIRDFGLE RVLIVDWDVH HGNGTQHIFE 720
721 SNPKVLYISL HRYEHGSFFP KGPDGNFDVV GKGAGRGFNV NIPWNKKGMG DLEYALAFQQ 780
781 LIMPIAYEFN PQLVLVSAGF DAAIGDPLGG CKVTAEGYGM LTHWLSALAS GRIIVCLEGG 840
841 YNVNSISYAM TMCTKTLLGD PVPTPQLGAT ALQKPPTVAF QSCVESLQQC LQVQRNHWRS 900
901 LEFVGRRLPR DPVVGENNNE DFLTASLRHL NISNDDATAA AGGLAGDRPD CGDERPSGSK 960
961 PKVKVKTLSD YLAENKEALE QSEMFAVYPL KTCPHLRLLR PEEAPRSLDS GAECSVCGST1020
1021 GENWVCLSCR HVACGRYVNA HMEQHSVEEQ HPLAMSTADL SVWCYACSAY VDHPRLYAYL1080
1081 NPLHEDKFQE PMAWTHGCAW REDGCYATGP DGRDEDDDDD NGAGSSICLR LERNN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
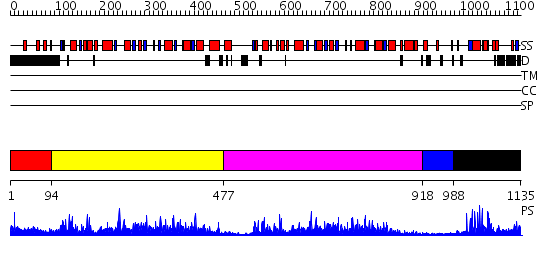
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [94..476] | 107.0 | No description for 2nvrA was found. | |
| 3 | View Details | [477..917] | 8.35 | No description for 2nvrA was found. | |
| 4 | View Details | [918..987] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [988..1135] | 22.0 | No description for 2g43A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)