













| General Information: |
|
| Name(s) found: |
FBpp0305870
[FlyBase]
sdt-PB / FBpp0088916 [FlyBase] |
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1292 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apicolateral plasma membrane
[IDA]
adherens junction [IDA] subapical complex [TAS] cell-cell adherens junction [NAS] cytoplasm [NAS] membrane [ISS][NAS] septate junction [TAS] apical cortex [IDA] plasma membrane [ISS] apical plasma membrane [IDA][NAS][TAS] |
| Biological Process: |
[NOT]establishment or maintenance of neuroblast polarity
[IEP][IMP]
cell-cell junction assembly [NAS] morphogenesis of an epithelium [NAS][TAS] zonula adherens assembly [IMP][TAS] establishment or maintenance of epithelial cell apical/basal polarity [IEP][IMP][IPI][TAS] establishment or maintenance of cell polarity [NAS] morphogenesis of a polarized epithelium [TAS] establishment or maintenance of polarity of embryonic epithelium [IMP] |
| Molecular Function: |
protein binding
[IPI][ISS][TAS]
guanylate kinase activity [ISS][NAS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLTMLSVNQD NGPHREMAVD CPDTFIARNK TPPRYPPPRP PQKHKKSTNT TTTTTITALT 60
61 NNDHANKMLI VAYHSSHQHE QLQQQHPSKT STTTTTIALD VATQNLYNQK QQNKLEQIEN 120
121 YENCLQSERN EQHEQQFEQQ KQHQATTAMA ATQVAQQQTP SHKLQATLSS DPNGNSNSNN 180
181 NSHIVGISSS SSSNNSSITD DFLCVVDGLY QGRKDTASPS SSAFDEVMSK HTLDSFGSIA 240
241 YRHLHQQHQA TSNGNSSSNT SNTNSNTNSN TNSNSNTNGN TSNNTAVSTK TATVTKTGVS 300
301 SSNSNSNSLN SSNSSMHTSS SSSGHSSNIA SATSSSSATS SSTVPDDLSL APPGYEVSQQ 360
361 QQQQHLVATP VTMLLPPMAK HRELPVDVPD SFIEMVKTTP RYPPPAHLSS RGSLLSNGSA 420
421 STAHTTLSSM GVAPSPVTAT AAAAASASAA CATTAVAAAA VSGVADGDAR RVADELNGNA 480
481 KPVPPPRDHL RVEKDGRLVN CSPAPQLPDR RAPGNASSGS SGATTHPLQH QQIAQIVEPT 540
541 LEQLDSIKKY QEQLRRRREE EERIAQQNEF LRNSLRGSRK LKALQDTATP GKAVAQQQQQ 600
601 ATLATQVVGV ENEAYLPDED QPQAEQIDGY GELIAALTRL QNQLSKSGLS TLAGRVSAAH 660
661 SVLASASVAH VLAARTAVLQ RRRSRVSGPL HHSSLGLQKD IVELLTQSNT AAAIELGNLL 720
721 TSHEMEGLLL AHDRIANHTD GTPSPTPTPT PAIGAATGST LSSPVAGPKR NLGMVVPPPV 780
781 VPPPLAQRGA MPLPRGESPP PVPMPPLATM PMSMPVNLPM SACFGTLNDQ NDNIRIIQIE 840
841 KSTEPLGATV RNEGEAVVIG RIVRGGAAEK SGLLHEGDEI LEVNGQELRG KTVNEVCALL 900
901 GAMQGTLTFL IVPAGSPPSV GVMGGTTGSQ LAGLGGAHRD TAVLHVRAHF DYDPEDDLYI 960
961 PCRELGISFQ KGDVLHVISR EDPNWWQAYR EGEEDQTLAG LIPSQSFQHQ RETMKLAIAE1020
1021 EAGLARSRGK DGSGSKGATL LCARKGRKKK KKASSEAGYP LYATTAPDET DPEEILTYEE1080
1081 VALYYPRATH KRPIVLIGPP NIGRHELRQR LMADSERFSA AVPHTSRARR EGEVPGVDYH1140
1141 FITRQAFEAD ILARRFVEHG EYEKAYYGTS LEAIRTVVAS GKICVLNLHP QSLKLLRASD1200
1201 LKPYVVLVAP PSLDKLRQKK LRNGEPFKEE ELKDIIATAR DMEARWGHLF DMIIINNDTE1260
1261 RAYHQLLAEI NSLEREPQWV PAQWVHNNRD ES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
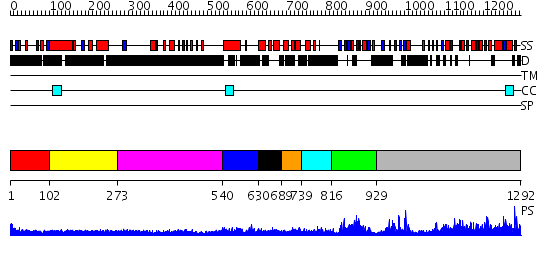
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [102..272] | 2.215995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [273..539] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [540..629] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [630..688] | 3.69897 | Hetero-tetrameric L27 (Lin-2, Lin-7) domain complexes as organization platforms of supra-molecular assemblies | |
| 6 | View Details | [689..738] | 2.77 | NMR structure of L27 heterodimer from C. elegans Lin-7 and H. sapiens Lin-2 scaffold proteins | |
| 7 | View Details | [739..815] | 5.522879 | NMR structure of L27 heterodimer from C. elegans Lin-7 and H. sapiens Lin-2 scaffold proteins | |
| 8 | View Details | [816..928] | 20.69897 | Solution structure of the PDZ domain of Pals1 protein | |
| 9 | View Details | [929..1292] | 89.221849 | Psd-95; Guanylate kinase-like domain of Psd-95 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)