













| General Information: |
|
| Name(s) found: |
Dok-PA /
FBpp0071008
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 622 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell cortex
[IDA]
|
| Biological Process: |
dorsal closure
[IMP]
imaginal disc fusion, thorax closure [IMP] stress-activated protein kinase signaling pathway [IMP] dorsal closure, elongation of leading edge cells [IMP] imaginal disc-derived wing morphogenesis [IMP] |
| Molecular Function: |
insulin receptor binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVEIPVLAG YLNVPTQTGF SLNRISKKKA CSRYCLLFKA SRHGIARLEM CESKEDRNPK 60
61 IFTLENCVKI TQEPPPERLI HIVKRQATLT LSTSSEEELK DWITALQTVA FCDTSPSGGI 120
121 GAIEEDNDLY CSSFDGLFII TLIPSEASIR CCIEPKTYML QMTPTELQLK SEDLGATIAM 180
181 WPYRFIRKYG YRDGKFTFEA GRKCTTGEGV FTLDHTNPQE VFRCMSAKMK SMKKLISGDS 240
241 LSTLECGENQ FSAAAGMEPG SRSPLPPSPS SNPHGGEFEI NSTQSCISLR GFISSNDSLN 300
301 NFSAGGGGAS GATATGGGVL GSTGGITLSV PANSNSNSSS SKHIPNKPPR KSPTTHCDKY 360
361 RNIVKYEPVA ITTISSPSAN SDVNSSVILK PLEGESISLP ETAADLPPDL PLRNESASKA 420
421 PPPERDYECI ENITEAWRTR GVGDVRHSER MPILCDTSEF VRQRSVSKST CGSQNTPKII 480
481 DIDIGEGCTS ATSISESNYD RLDFLSPNNK TSSGYKTIVN VTPGNRIRCS PPPPNEYELI 540
541 ASPDTESFRK ADDSHRGYGV LRKASTTTNN NNLGISNSNI SGGSSTITTA TANKGGNLEA 600
601 AAIDHRSYNG LNYTMVSKPK RV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
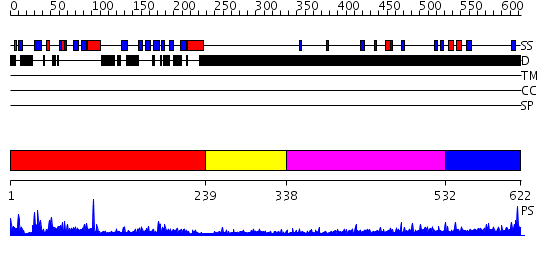
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..238] | 60.522879 | Insulin receptor substrate 1, IRS-1 | |
| 2 | View Details | [239..337] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [338..531] | 7.215997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [532..622] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)